Morphological, molecular and 3D synchrotron X-ray tomographic characterizations of Helicascus satunensis sp. nov., a novel mangrove fungus
- Published
- Accepted
- Received
- Academic Editor
- Tony Robillard
- Copyright
- © 2024 Preedanon et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2024. Morphological, molecular and 3D synchrotron X-ray tomographic characterizations of Helicascus satunensis sp. nov., a novel mangrove fungus. PeerJ 12:e18341 https://doi.org/10.7717/peerj.18341
Abstract
A new species of Helicascus satunensis sp. nov. was collected on mature dead fruits of the Nypa palm in Satun Province, southern Thailand. Its morphological characteristics are similar to those of the genus Helicascus. Recently, a genus Helicascus with three species from marine habitats worldwide was studied. The morphology of this fungus was investigated and combined with multigene sequence analyzes of small subunit (SSU), large subunit (LSU), internal transcribed spacer (ITS) ribosomal DNA, translation elongation factor 1-alpha (TEF-1α) and RNA polymerase II (RPB2) genes. Morphologically, H. satunensis sp. nov. is characterized by semi-immersed, lenticular ascomata, multilocules, a bitunicate ascus and smooth, obovoid, dark brown ascospores that are one-septate and unequally two-celled. In addition, 3D visualization using synchrotron X-ray tomography was performed to investigate the interaction between fruiting body and substrata. Molecular phylogeny with multigene revealed that H. satunensis sp. nov. belongs to the family Morosphaeriaceae, order Pleosporales, class Dothideomycetes. Furthermore, H. satunensis sp. nov. forms a well-supported clade with Helicascus species described from marine habitats. Based on the unique morphological and molecular evidence, we propose this fungus, H. satunensis sp. nov., as a new species for Helicascus.
Introduction
Kohlmeyer (1969) described the distinct marine ascomycete Helicascus obtained from dead prop roots of the mangrove Rhizophora mangle. A type species, Helicascus kanaloanus, is characterized by an immersed ascostroma composed of multilocules that share a common periphysate ostiole lying under pseudostromatic tissues. The asci are subcylindrical bitunicate and pediculated, and the endoascus is usually coiled at the base. The ascospores are brown to dark brown at maturity and are frequently asymmetrically two-celled with a mucilaginous sheath in this species (Kohlmeyer, 1969).
Since 1991, a number of Helicascus-like species have been described from freshwater and marine habitats based on their common morphological characteristics and DNA sequences (Hyde, 1991; Zhang et al., 2013; Zhang et al., 2014; Zhang et al., 2015; Luo et al., 2016; Preedanon et al., 2017; Zhang et al., 2024). However, two new genera, Aquihelicascus and Neohelicascus, were excluded from the genus Helicascus due to their morphological and multigene phylogeny (Dong et al., 2020). Aquihelicascus was established to accommodate one new combination and two new species. Neohelicascus was introduced to accommodate one new species and seven new combinations (Dong et al., 2020).
Recently, three marine species in the genus Helicascus were identified, H. kanaloanus, H. nypae and H. mangrovei, based on morphological and molecular data (Kohlmeyer, 1969; Hyde, 1991; Preedanon et al., 2017). Helicascus nypae was found on Nypa palm fronds from Southeast Asia. It is characterized as having ascomata with immersed multilocules with a single common central ostiole, bitunicate asci with a long, narrow and coiled endoascus, and unequally two-celled, verrucose ascospores surrounded by a gelatinous sheath (Hyde, 1991). Subsequently, Preedanon et al. (2017) reported H. mangrovei obtained from decaying mangrove wood in Thailand. The unique morphological characteristics of H. mangrovei include multilocular ascomata semi-immersed under a thick clypeus that forms pseudostromata, clavate pedicellate asci in a hamathecium of cellular pseudoparaphyses, dark brown color at maturity, and unequally two-celled ascospores with one apiculate end.
The aim of this study is to report on novel ascomycete found in Thai mangrove habitats. The microscopic morphology of the fungal fruiting bodies and host tissues was visualized in three dimensions using synchrotron radiation X-ray tomography, which enables high-resolution and non-invasive visualization of internal features without the need for serial sections and staining reagents. This capability is simply unattainable with conventional characterization tools (Friis et al., 2014; Sena et al., 2022; Becher, Sheppard & Grunwaldt, 2023). Furthermore, we provide the molecular phylogeny of the combined SSU, LSU, ITS rDNA, TEF-1α and RPB2 sequences to confirm their taxonomic position.
Materials and Methods
Sample collection, isolation, morphological examination, and materials availability
Mature dead Nypa palm fruits were collected from mangroves at Mangrove Forest Resource Development Station 36 in Satun Province, southern Thailand (6°54′14.9616″N and 99°41′17.4912″E). Collecting procedure was made as previously described in Preedanon et al. (2017). The collected Nypa palm fruits were placed in a sealed plastic Ziploc bag and brought back to the laboratory for further examination. The specimens were washed with natural seawater in order to remove sediment and other debris then kept in a moist plastic box and incubated at room temperature (approximately 25–28 °C). The samples were examined directly under a stereo-zoom microscope for the presence of H. satunensis sp. nov. Photographic documentation of the sporulating structures was carried out using the Olympus BX51 and Olympus DP21 software (Olympus, Tokyo). The ascomata specimens were fixed with embedding matrix on stage and cutting sections through a cryostat microtome (MEV; SLEE Mainz, Mainz, Germany). The fresh ascomata of H. satunensis sp. nov. were selected for single spore isolation (Vrijmoed, 2000). Spore suspension was diluted and plated on 1.5% seawater corn meal agar (SCMA) medium with the addition of antibiotics (streptomycin sulfate 0.5 g/L, penicillin G 0.5 g/L). The germinated spores were placed onto freshly SCMA medium and incubated at room temperature (approximately 25–28 °C) (Preedanon et al., 2017). Axenic cultures (BCC 83546, BCC 86189, BCC 86190) were then transferred to 1.5% seawater potato dextrose agar (SPDA). Colony characteristics, growth and sporulation were observed and recorded. The type cultures were deposited at the BIOTEC Culture Collection (BCC), Pathum Thani, Thailand. In addition, dried voucher type specimens (BBH 50658, BBH 50659 and BBH 50660) were deposited at BIOTEC Bangkok Herbarium (BBH), Pathum Thani, Thailand.
Three-dimensional synchrotron X-ray tomography
The microstructure of the fungal fruiting bodies and the outer exocarp of Nypa palm fruit was visualized in three dimensions using synchrotron radiation X-ray tomographic microscopy (SRXTM). Prior to the SRXTM experiment, the fresh fungal fruiting bodies samples were fixed with 3% formaldehyde. For tomographic imaging, each sample was placed in a sample holder attached to a brass stub with glue to stabilize the sample during tomography scanning. The SRXTM examination of the samples was carried out at the X-ray tomographic microscopy beamline (BL1.2W: XTM) at the Siam Photon Source Facility, Synchrotron Light Research Institute. The X-ray beam was generated from a 2.2-Tesla multipole wiggler radiation source optimized with a toroidal focusing mirror and filtered with aluminum foils to achieve an average energy of 10 keV. All X-ray projections were acquired with a pixel size of 3.61 µm using an imaging system consisting of a 2X objective lens-coupled microscope (Optique Peter, Lentilly, France), a YAG-Ce scintillator (CRYTUR, Turnov, Czech Republic) and the PCO.edge 5.5 sCMOS camera (Excelitas PCO GmbH, Kelheim, Germany). To enhance fine details of the entire sample, a tomographic volume was reconstructed from enlarged composite projections obtained from multiple scans. Each scan covered 180°with a step of 0.2°, resulting in a dataset. Subsequently, the X-ray projection datasets underwent pre-processing, which included flat-field correction, beam intensity normalization and image stitching. Tomographic reconstruction was performed using Octopus Reconstruction software (TESCAN, Ghent, Belgium). The resulting computed tomographic slices were analyzed using ImageJ (http://rsbweb.nih.gov/ij/) and Fiji (http://fiji.sc/Fiji), and the 3D visualization of the tomographic volume was displayed using Drishti software (Limaye, 2012).
DNA extraction, PCR amplification and DNA sequencing
Genomic DNA from fungal mycelia was extracted according to the methods of O’Donnell et al. (1997) and Sakayaroj, Pang & Jones (2011). Ribosomal DNA genes (ITS, SSU, LSU) and protein-coding gene sequences (TEF-1α, RPB2) were amplified by polymerase chain reaction (PCR). The ITS rDNA region was amplified with the primer pair ITS4/ITS5 (White et al., 1990), the SSU region with NS1/NS4 (White et al., 1990), the LSU region with LROR/LR5 (Vilgalys & Hester, 1990), the TEF1-α region with EF1-983F/EF1-2218R (Rehner & Buckley, 2005), the RPB2 region with fRPB2-5F/fRPB2-7cR (Liu, Whelen & Hall, 1999) (Table 1). The component of PCR reaction was performed in a total volume of 50 µL, containing 1 µL DNA template (30–50 ng/ µL), 1 µL of each forward and reverse primers (10 µM), 10 µL master mix of Taq DNA polymerase (Thermo Fisher Scientific Inc., Waltham, MA, USA) and 37 µL of double-distilled water. The PCR conditions for all the genes used were set up using the T100TM thermal cycler (BIO-RAD Laboratories, Inc., California) (Table 1). The PCR products were subsequently purified and sequenced by Macrogen (Seoul, South Korea).
| DNA region | Primer name | Amplification profile | Reference | ||
|---|---|---|---|---|---|
| Denaturation | Repeat steps | Extension | |||
| Internal transcribed spacer rDNA (ITS) | ITS5 ITS4 | 94 °C (2 min) | 35 cycles 94 °C (1 min) 54 °C (1 min) 72 °C (2 min) |
72 °C (10 min) | White et al. (1990) |
| 18S small subunit rDNA (SSU) | NS1 NS4 | 94 °C (2 min) |
35 cycles 94 °C (1 min) 55 °C (1 min) 72 °C (2 min) |
72 °C (10 min) |
White et al. (1990) |
| 28S large subunit rDNA (LSU) | LROR LR5 | 94 °C (2 min) |
35 cycles 94 °C (1 min) 55 °C (1.5 min) 72 °C (2.5 min) |
72 °C (10 min) |
Vilgalys & Hester (1990) |
| Translation elongation factor 1-alpha (TEF 1-α) | EF1-983F EF1-2218R | 95 °C (2 min) |
35 cycles 95 °C (1 min) 54 °C (1 min) 72 °C (2 min) |
72 °C (10 min) |
Rehner & Buckley (2005) |
| RNA polymerase II second largest subunit (RPB2) | fRPB2-5F fRPB2-7cR | 94 °C (3 min) |
35 cycles 94 °C (1 min) 54 °C (1 min) 72 °C (1.5 min) |
72 °C (8 min) |
Liu, Whelen & Hall (1999) |
| Taxon | Strain | GenBank accession no. | ||||
|---|---|---|---|---|---|---|
| LSU rDNA |
SSU rDNA |
ITS rDNA |
TEF-1α | RPB2 | ||
| Aquihelicascus songkhlaensis | MFLUCC 18-1154T | MN913692 | – | MT627680 | MT954380 | – |
| Aquihelicascus songkhlaensis | MFLUCC 18-1273 | MN913724 | MT864319 | MT627696 | MT954369 | MT878464 |
| Aquihelicascus songkhlaensis | MFLUCC 18-1278 | MN913726 | MT864318 | MT627693 | MT954366 | MT878458 |
| Aquihelicascus thalassioideus | MFLUCC 10-0911T | KC886636 | KC886637 | KC886635 | – | – |
| Aquihelicascus thalassioideus | MJF 14020-2 | KP637165 | – | KP637162 | – | – |
| Aquihelicascus thalassioideus | JCM 17526 | AB807558 | AB797268 | LC014554 | AB808534 | – |
| Aquihelicascus thalassioideus | CBS 110441 | AB807557 | AB797267 | LC014553 | AB808533 | – |
| Aquihelicascus thalassioideus | KUMCC 19-0094 | MT627668 | – | MT627689 | – | – |
| Aquihelicascus yunnanensis | MFLUCC 18-1025T | MN913711 | MT864292 | MT627728 | MT954391 | – |
| Aquilomyces patris | CBS 135661T | KP184041 | KP184077 | KP184002 | – | – |
| Aquilomyces patris | CBS 135760 | KP184042 | KP184078 | KP184004 | – | – |
| Aquilomyces patris | CBS 135662 | KP184043 | KP184079 | KP184003 | – | – |
| Aquilomyces rebunensis | CBS 139684T | AB807542 | AB797252 | AB809630 | AB808518 | – |
| Clypeoloculus akitaensis | CBS 139681T | AB807543 | AB797253 | AB809631 | AB808519 | – |
| Clypeoloculus hirosakiensis | CBS 139682T | AB807550 | AB797260 | AB809638 | AB808526 | – |
| Clypeoloculus microsporus | CBS 139683T | AB807535 | AB797245 | AB811451 | AB808510 | – |
| Clypeoloculus towadaensis | CBS 139685T | AB807549 | AB797259 | AB809637 | AB808525 | – |
| Didymella fucicola | JK 2932 | EF177852 | – | EF192138 | – | – |
| Falciformispora lignatilis | BCC 21118 | GU371827 | GU371835 | – | GU371820 | – |
| Halojulella avicenniae | BCC 18422 | GU371823 | GU371831 | – | GU371816 | GU371787 |
| Halojulella avicenniae | BCC 20173 | GU371822 | GU371830 | – | GU371815 | GU371786 |
| Halojulella avicenniae | JK 5326A | GU479790 | GU479756 | – | – | – |
| Helicascus kanaloanus | A 237 | – | AF053729 | – | – | – |
| Helicascus kanaloanus | ATCC 18591 | KX639748 | KX639744 | KX957961 | KX639756 | KX639752 |
| Helicascus mangrovei | BCC 68258T | KX639745 | KX639741 | KX957958 | KX639753 | KX639749 |
| Helicascus mangrovei | BCC 68260 | KX639746 | KX639742 | KX957959 | KX639754 | KX639750 |
| Helicascus mangrovei | BCC 74471 | KX639747 | KX639743 | KX957960 | KX639755 | KX639751 |
| Helicascus nypae | BCC 36751 | GU479788 | GU479754 | – | GU479854 | GU479826 |
| Helicascus nypae | BCC 36752 | GU479789 | GU479755 | – | GU479855 | GU479827 |
| Helicascus satunensis | BCC 83546T | PP866393 | PP873998 | PP873995 | PP915719 | PP915722 |
| Helicascus satunensis | BCC 86189 | PP866394 | PP873999 | PP873996 | PP915720 | - |
| Helicascus satunensis | BCC 86190 | PP866395 | PP874000 | PP873997 | PP915721 | PP915723 |
| Leptosphaeria maculans | AFTOL ID-277 | DQ470946 | DQ470993 | – | DQ471062 | DQ470894 |
| Massarina igniaria | CBS 845.96 | DQ810223 | DQ813511 | – | – | – |
| Microvesuvius unicellularis | AD 291626 | OQ799383 | – | OQ799384 | OQ866586 | – |
| Microvesuvius unicellularis | AD 291633T | OQ799391 | – | OQ799382 | OQ866585 | – |
| Montagnula opulenta | AFTOL ID-1734 | DQ678086 | AF164370 | – | – | DQ677984 |
| Morosphaeria muthupetensis | PUFD 87T | MF614796 | MF614797 | MF614795 | MF614798 | – |
| Morosphaeria ramunculicola | BCC 18404 | GQ925853 | GQ925838 | – | – | – |
| Morosphaeria ramunculicola | BCC 18405 | GQ925854 | GQ925839 | – | – | – |
| Morosphaeria ramunculicola | JK 5304B | GU479794 | GU479760 | – | – | GU479831 |
| Morosphaeria ramunculicola | KH 220 | AB807554 | AB797264 | – | AB808530 | – |
| Morosphaeria velatispora | BCC 17059 | GQ925852 | GQ925841 | – | – | – |
| Morosphaeria velatispora | NBRC 107812 | AB807556 | AB797266 | LC014572 | AB808532 | – |
| Neohelicascus aegyptiacus | MFLU 12-0060T | KC894853 | KC894852 | – | – | – |
| Neohelicascus aquaticus | KUMCC 19-0107 | MT627662 | MT864314 | MT627719 | MT954384 | – |
| Neohelicascus aquaticus | KUMCC 17-0145 | MG356477 | MG356487 | MG356479 | MG372317 | – |
| Neohelicascus aquaticus | MFLUCC 17-2300 | MG356478 | – | MG356480 | MG372316 | – |
| Neohelicascus aquaticus | MFLUCC 10-0918T | KC886640 | KC886638 | KC886639 | MT954384 | – |
| Neohelicascus aquaticus | MAFF 243866 | AB807532 | AB797242 | AB809627 | AB808507 | – |
| Neohelicascus chiangraiensis | MFLUCC 13-0883T | KU900585 | KU900587 | KU900583 | KX455849 | – |
| Neohelicascus elaterascus | MAFF 243867 | AB807533 | AB797243 | AB809626 | AB808508 | – |
| Neohelicascus elaterascus | CBS 139689 | LC014608 | LC014603 | LC014552 | LC014613 | – |
| Neohelicascus elaterascus | MFLUCC 18-0985 | MT627658 | MT864335 | MT627735 | – | – |
| Neohelicascus elaterascus | MFLUCC 18-0993 | MT627659 | MT864333 | MT627730 | – | – |
| Neohelicascus elaterascus | HKUCC 7769 | AY787934 | AF053727 | – | – | – |
| Neohelicascus gallicus | BJFUCC 200228 | KM924831 | – | KM924833 | – | – |
| Neohelicascus gallicus | CBS 123118 | KM924832 | – | – | – | – |
| Neohelicascus gallicus | BJFUCC 200224 | KM924830 | – | – | – | – |
| Neohelicascus griseofavus | MFLUCC 16-0869T | OP377964 | OP378041 | OP377878 | OP473055 | – |
| Neohelicascus submersus | MFLU 20-0436T | MT627656 | MT864340 | MT627742 | – | – |
| Neohelicascus unilocularis | MJF 14020T | KP637166 | – | KP637163 | – | – |
| Neohelicascus unilocularis | MJF 14020-1 | KP637167 | – | KP637164 | – | – |
| Neohelicascus uniseptatus | MFLUCC 15-0057T | KU900584 | – | KU900582 | KX455850 | – |
| Paradendryphiella arenariae | AFTOL ID-995T | DQ470971 | DQ471022 | – | DQ677890 | DQ470924 |
| Parastagonospora avenae | AFTOL ID-280 | AY544684 | AY544725 | – | DQ677885 | DQ677941 |
| Phaeodothis winteri | AFTOL ID-1590 | DQ678073 | DQ678021 | – | DQ677917 | DQ677970 |
| Phaeosphaeria eustoma | AFTOL ID-1570 | DQ678063 | DQ678011 | – | DQ677906 | DQ677959 |
| Platychora ulmi | CBS 361.52 | EF114702 | EF114726 | – | – | – |
| Plenodomus biglobosus | CBS 303.51 | GU301826 | – | – | GU349010 | – |
| Setoseptoria arundinacea | CBS 619.86 | DQ813509 | DQ813513 | – | – | – |
| Stemphylium vesicarium | CBS 191.86T | DQ247804 | DQ247812 | – | DQ471090 | DQ247794 |
| Trematosphaeria pertusa | CBS 122371 | FJ201992 | FJ201993 | – | – | – |
| Outgroup | ||||||
| Lophiostoma macrostomum | JCM 13544 | AB619010 | AB618691 | JN942961 | LC001751 | JN993491 |
| Sigarispora arundinis | JCM 13550 | AB618998 | AB618679 | JN942964 | LC001737 | JN993482 |
Notes:
- T
-
Ex-type strain
Phylogenetic analyses
Multiple sequence alignments were analyzed with the closely matched sequences obtained from GenBank (Table 2) according to Jones et al. (2015), Maharachchikumbura et al. (2016) and Hongsanan et al. (2017), Dong et al. (2020), Yang et al. (2023a) and Yang et al. (2023b). The newly generated sequences from this study are listed in Table 2. The nucleotide sequences were assembled and aligned using BioEdit 7.2.5 (Hall, 1999) and Muscle 3.8.31 (Edgar, 2004). Specifically, NCBI blast searches were used to determine sequence similarity to sequences published in the GenBank database. Phylogenetic analyses of the combined SSU, LSU, ITS rDNA, TEF-1α and RPB2 sequences were performed using maximum likelihood (ML) and Bayesian algorithms. Maximum likelihood (ML) analysis was evaluated in RAxMLHPC2 on XSEDE (Stamatakis, 2014) via the CIPRES Science Gateway platform (Miller, Pfeiffer & Schwartz, 2010) under the GTR + GAMMA model with BFGS method to optimize the GTR rate parameters. Finally, Bayesian posterior probabilities of branches were performed using MrBayes 3.2.6 (Ronquist et al., 2012), with the best-fitting model (GTR+I+G) selected by AIC in MrModeltest 2.2 (Nylander, 2004), which was tested with hierarchical likelihood ratios (hLRTs). Three million generations were run in four Markov chains and a sample was drawn every 100 generations with a burn-in value of 3,000 sampled trees. Finally, the consensus tree was displayed using the interactive Tree Of Life (iTOL) (Letunic & Bork, 2021) and adjusted in Adobe Photoshop 2020. All sequences obtained in this study were submitted to GenBank, and the typification were published in the MycoBank database (Crous et al., 2004). The resulting alignments were submitted to TreeBASE (submission numbers: 31389).
Nomenclature
The electronic version of this article in Portable Document Format (PDF) will represent a published work according to the International Code of Nomenclature for algae, fungi, and plants, and hence the new names contained in the electronic version are effectively published under that Code from the electronic edition alone, so there is no longer any need to provide printed copies. In addition, new names contained in this work have been submitted to MycoBank from where they will be made available to the Global Names Index. The unique MycoBank number can be resolved and the associated information viewed through any standard web browser by appending the MycoBank number contained in this publication (MB 854336) to the prefix http://www.mycobank.org/MB/. The online version of this work is archived and available from the following digital repositories: PeerJ, PubMed Central SCIE, and CLOCKSS.
Results
Taxonomy
Type: THAILAND, Satun Province, mangrove forests, on a piece of dead palm (Nypa fruticans) fruit, 22 December 2016, S. Preedanon, A. Klaysuban, O. Pracharoen & J. Sakayaroj, BBH 50658, holotypus, cultura dessicata, (holotype designated here) (BIOTEC Bangkok Herbarium, Pathum Thani, Thailand).
Ex-type culture: MCR 00699 (BCC 83546) (BIOTEC Culture Collection, Pathum Thani, Thailand).
Etymology: ‘satunensis’ referring to the collecting location, Satun Province, southern Thailand, where the fungus was collected.
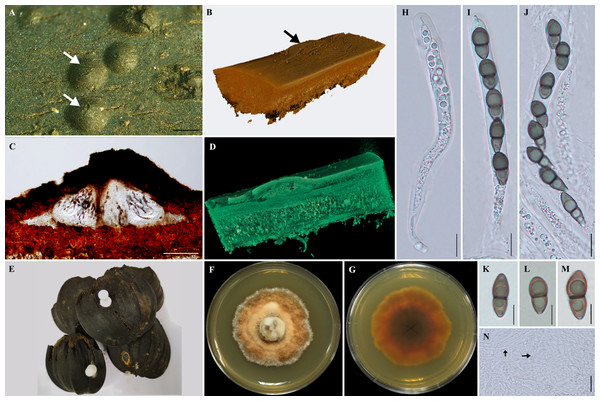
Sexual morph: Ascomata 1,000–2, 400 × 160–280 µm, semi-immersed, lenticular ascomata, 3–4 locules, dark brown to black, carbonaceous, solitary (Fig. 1).
Three-dimensional synchrotron X-ray tomographic analysis reveals that the fungal tissues growing in the outer exocarp of Nypa palm fruits, enclosing 3–4 locules with flattened base, horizontally arranged under the pseudostroma. Cellular, numerous, persistent, hyaline pseudoparaphyses.
Asci 475–642. 5 × 62.5–80 µm, 8-spored, bitunicate asci, cylindrical, thick-walled, short hook pedunculate, with an ocular chamber. Ascospores 22.5–25 × 5–8.75 µm, unequally two-celled, smooth, dark-brown, and slightly constricted at the septum, thick-walled (Fig. 1).
Habitat and distribution: mangrove forests, Satun Province, southern Thailand.
Asexual morph: Undetermined
Culture characteristics: Ascospores germinated on SCMA after 1–2 days, colonial grown on SPDA attaining 2–3 cm in diameter after 60 days incubation at room temperature (approximately 25–28 °C), dense, circular, irregular and grey (7D1) with orange patches (7C5) in the center, white (7A1) at the edge; dark brown at reverse side. Colour codes in the fungal description follow “Methuen Handbook of Colour” (Kornerup & Wanscher, 1978).
Phylogenetic analyses
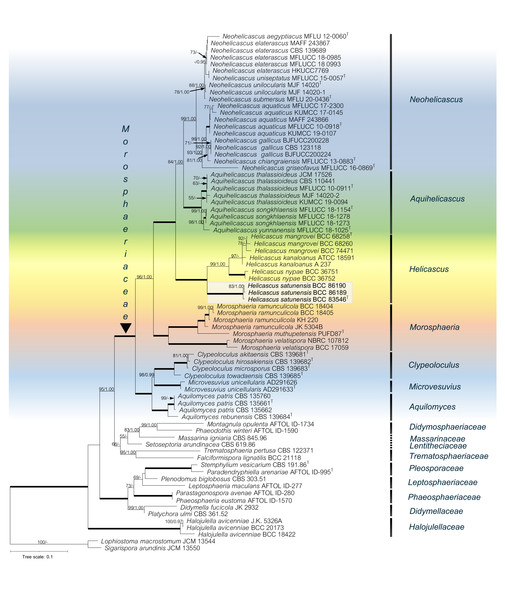
The phylogenetic relationships of the Pleosporales, Dothideomycetes were reconstructed using the combined five-gene dataset (SSU, LSU, ITS rDNA, TEF-1α, RPB2), with Lophiostoma macrostomum JCM 13544 and Sigarispora arundinis JCM 13550 as the outgroups (Table 2). The alignment of 74 taxa comprised 5,844 base pairs (1,342 for SSU, 1,358 for LSU, 1,192 for ITS, 968 for TEF-1α and 984 for RPB2). Total 3,825 characters were constant; 1,570 characters were parsimony-informative and 449 variable characters were parsimony-uninformative. Phylogenetic analyses showed that our novel species (in bold) belongs to the Morosphaeriaceae. Although the topology of the BI tree and the exhibited ML tree are comparable, the BI tree is not shown. The phylogenetic trees representing the unique position of other species in the marine habitat were deposited in MycoBank. Significant ML bootstrap values (≥50%) and Bayesian posterior probabilities (≥0.95) are indicated in the phylogenetic tree (Fig. 2).
Figure 1: Morphological features of Helicascus satunensis sp. nov.
(A) Carbonaceous ascomata on the exocarp of Nypa palm fruit. (B) The 3D visualization of the ascoma by X-ray tomography (arrow). (C) Vertical section through an ascoma. (D) Section through an ascoma using 3D visualization. (E) Dead Nypa palm fruits. (F–G) Obverse and reverse views of cultures grown on SPDA after 60 days. (H–J) Subcylindrical bitunicate asci. (K–M) Ascospores. (N) Pseudoparaphyses (arrow). Scale bars A = 1 mm, C =300 µm, H–J = 100 µm, K–M = 10 µm, N = 20 µm.Figure 2: The RAxML phylogenetic tree of Helicascus satunensis (BCC 83546, BCC 86189, BCC 86190) resulted from the combined of LSU, SSU, ITS, TEF-1α and RPB2 sequences.
Lophiostoma macrostomum and Sigarispora arundinis were used as outgroups. Maximum likelihood (BSML, left) equal to or greater than 50% are shown above each branch. Bayesian posterior probabilities (BYPP, right) equal to or greater than 0.95 are shown below each branch. The nodes that are strongly supported by bootstrap proportions (100%) and posterior probabilities (1.00) are shown in a thicker line. Abbreviations: T = ex-type. Novel species is demonstrated in bold.In the multigene phylogenetic analysis, the Morosphaeriaceae are divided into subclades representing the seven accepted genera including Neohelicascus, Aquihelicascus, Helicascus, Morosphaeria, Clypeoloculus, Microvesuvius, and Aquilomyces. Our fungal strains (BCC 83546, BCC 86189 and BCC 86190) are monophyletic and well placed in the Morosphaeriaceae with robust bootstrap and Bayesian supports. They are phylogenetically distinct from the type species H. kanaloanus and form sister subclades with H. nypae and H. mangrovei (Fig. 2). Within the Helicascus subclade, we compared the base substitutions of our new fungus with the type species H. kanaloanus. The result shows the base substitutions at several sites of SSU (960/969 = 99.0% similarity), LSU (803/853 = 94.1% similarity), ITS rDNA (448/714 = 62.7% similarity), TEF-1 α (872/933 = 93.4% similarity) and RPB2 (793/899 = 88.2% similarity) (Table 3).
Discussion
Taxonomy
Jones et al. (2022) reported 1,900 marine fungi in 769 genera that have evolved for marine life as saprobes, parasites and endophytes. Devadatha et al. (2021) and Zhang et al. (2024) reported that many new taxa have been described from mangrove trees and salt marsh plants. Among these host plants, palms found in mangroves and estuaries, such as Phoenix paludosa, Oncosperma tigillarium, and N. fruticans, harbour a great diversity of fungi (Zhang et al., 2024).
Some of the fungi are found as saprobes on the petioles of N. fruticans: Bacusphaeria nypae, Manglicola guatemaelensis, and Tirisporella baccariana (Jones et al., 1996; Suetrong et al., 2009; Abdel-Wahab et al., 2017). Acuminatispora palmarum, Fasciatispora nypae, Helicascus nypae, Neomorosphaeria mangrovei, Pleurophomopsis nypae, Striatiguttula nypae, and S . phoenicis grow on submerged rachis and petioles of N. fruticans and Ph. paludosa (Hyde et al., 1999; Hyde & Alias, 2000; Loilong et al., 2012; Zhang et al., 2018; Zhang et al., 2019; Zhang et al., 2024). A few fungi, however, were discovered on Nypa fruits: Anthostomella nypae., Fasciatispora spp., and Vaginatispora nypae (Jayasiri et al., 2019; Zhang et al., 2024).
| DNA sequence | Number of bases for comparison (bp) |
Helicascus kanaloanus | H. nypae | H. mangrovei | |||
|---|---|---|---|---|---|---|---|
| Base substitutions | % similarity |
Base substitutions | % similarity |
Base substitutions | % Similarity |
||
| SSU rDNA | 969 | 9 | 99.0 | 14 | 98.5 | 9 | 99.0 |
| LSU rDNA | 853 | 50 | 94.1 | 47 | 94.5 | 40 | 95.3 |
| ITS rDNA | 714 | 266 | 62.7 | ND | ND | 265 | 62.9 |
| TEF-1α | 933 | 61 | 93.4 | 74 | 92.0 | 64 | 93.1 |
| RPB2 | 899 | 106 | 88.2 | 115 | 87.2 | 107 | 88.1 |
Notes:
- ND
-
Not determined
The genus Helicascus is a distinct marine ascomycete characterized by having a pseudostroma composed of host cells enclosed in fungal hyphae, subcylindrical asci, uniseriate, obovoid, dark brown color at maturity, and asymmetrical ascospores (Kohlmeyer, 1969). Recently, three species have been identified in the genus, namely, H. kanaloanus (type species), H. nypae, and H. mangrovei (Kohlmeyer, 1969; Hyde, 1991; Preedanon et al., 2017). We found a new fungus, H. satunensis, that inhabits the brackish waters of Nypa palm fruit in Satun Province, southern Thailand. Helicascus satunensis shares similar ascostromata with H. kanaloanus and H. nypae in having semi-immersed or immersed, carbonaceous, multilocules in the ascostromata arranged under a black pseudoclypeus, while H. mangrovei does not have separate locules in the ascomata (Table 4).
| Helicascuskanaloanus | H. nypae | H. mangrovei | H. satunensis sp. nov. | |
|---|---|---|---|---|
| Pseudostromata | ||||
| Size (µm) | 600–780 × 1,250–2,750 | 260–390 × 750–1,500 | 1,500–1, 750 × 1,500–2,500 | 1,000–2, 400 × 160–280 |
| Position on substrata | Immersed | Immersed | Semi-immersed | Semi-immersed |
| Locules | Multilocules (3–4 loculi) | Multilocules (3–4 loculi) | Single locule | Multilocules (3–4 loculi) |
| Structure | Ampulliform, lenticular, horizontally arranged under a black pseudoclypeus | Lenticular, black, carbonaceous | Lenticular, flattened, carbonaceous, solitary, a locule covered by a pseudoclypeus | Lenticular, black, carbonaceous, solitary |
| Asci | ||||
| Size (µm) | 200–260 × 15–25 | 192–280 × 14–20 | 400–412. 5 × 25–30 | 475–642. 5 × 62.5–80 |
| Shape | Subcylindrical to oblong clavate, persistent, pedunculate, thick-walled | Subcylindrical, pedunculate | Subcylindrical, pedunculate, thick-walled | Cylindrical, thick-walled |
| Endoascus | With an apical apparatus coiling |
With an ocular chamber | With an apical apparatus coiling |
Short hook pedunculate with an ocular chamber |
| Pseudoparaphyses | Cellular, numerous, persistent | Cellular, numerous, persistent, anastomosing in a gel | Cellular, numerous, trabeculate, hyaline | Cellular, numerous, persistent, hyaline |
| Ascospores | ||||
| Size (µm) | 30–55 × 17–25 | 25–35 × 12 –15 | 40–45 × 18.5–20 | 22.5–25 × 5–8.75 |
| Sheath | Present in some collection | Present | Absent | Absent |
| Shape | Obovoid, brown, biseriate one-septate, constricted at the septum, dark-brown at maturity, unequally two-celled |
Uniseriate, obovoid, constricted at the septum, brown, sometimes at one or both ends apiculate, unequally two-celled |
Uniseriate, obovoid, unequally two-celled, slightly constricted at the septum, thick-walled and only one apiculate end, dark brown at maturity |
Constricted at the septum, thick-walled, unequally two-celled, dark brown at maturity |
| Ornamentation | Smooth wall | Verrucose wall | Smooth wall | Smooth wall |
| Host | Dead mangrove wood | Nypa fruticans, Phoenix paludosa fronds | Dead mangrove wood | Nypa fruticans fruit |
| Asexual morph | Undetermined | Pleurophomopsis nypae | Undetermined | Undetermined |
Pseudostroma is a unique taxonomic characteristic of the Morosphaeriaceae at the genus level. Zhang et al. (2015) reported that multilocular pseudostroma are important in delineating species of Helicascus-like species. Members of the Morosphaeriaceae develop somatic hyphae into ascostroma, which subsequently form locules that include the genera Helicascus, Neohelicascus, Aquihelicascus, Morosphaeria, Clypeoloculus, and Aquilomyces (Dong et al., 2020). The coiling and stretching mechanism of the basal endoascus with an ocular chamber are regarded as unique types of asci in the genus Helicascus. Our new fungus H. satunensis shares this type of asci with three other species in the genus (Zhang et al., 2015). All species share the same arrangement of cellular pseudoparaphyses. The ascospores of H. satunensis can be distinguished from those of other species because they are smaller (22.5–25 × 5–8.75 µm) than those of H. kanaloanus (30–55 × 17–25 µm), H. nypae (25–35 × 12–15 µm), and H. mangrovei (40–45 × 18.5–20 µm). Germ pores were not observed in H. satunensis, while they appeared at only one end in H. mangrovei (Preedanon et al., 2017). Moreover, the unequal number of H. satunensis 2-cell cones with constriction of ascospores could be a defined taxonomic marker at the species level in the genus Helicascus.
Molecular phylogeny
Phylogenetic analyses of multigene sequences revealed that Helicascus satunensis forms a well-supported clade within the Morosphaeriaceae, Pleosporales, Dothideomycetes. The Morosphaeriaceae family was established by Suetrong et al. (2009) based on morphological features and strong phylogenetic support. Currently, the family comprises eight genera: Aquihelicascus (Dong et al., 2020), Aquilomyces (Knapp et al., 2015), Clypeoloculus (Tanaka et al., 2015), Helicascus (Kohlmeyer, 1969; Dong et al., 2020), Microvesuvius (Fryar, Réblová & Catcheside, 2023), Morosphaeria (Suetrong et al., 2009), Neohelicascus (Dong et al., 2020), and Neomorosphaeria (Zhang et al., 2024).
Members in the Morosphaeriaceae family are found on submerged dead twigs in freshwater and marine environments. The multigene phylogeny comprising freshwater taxa formed sister clades to the marine fungal lineages. Two new freshwater genera, Aquihelicascus and Neohelicascus, were excluded from the genus Helicascus due to morphological and molecular evidence (Dong et al., 2020). Aquihelicascus was established to accommodate one new combination (A. thalassioideus) and two new species (A. songkhlaensis and A. yunnanensis). Neohelicascus was introduced to accommodate one new species (N. submersus) and seven new combinations (N. elaterascus, N. chiangraiensis, N. unilocularis, N. uniseptatus, N. aegyptiacus, N. gallicus, and N. aquaticus) (Dong et al., 2020). The genera Morosphaeria (M. ramunculicola, M. muthupetensis, M. velatispora) (Suetrong et al., 2009; Devadatha et al., 2018), Neomorosphaeria (Zhang et al., 2024), and Helicascus (H. kanaloanus, H. nypae, H. mangrovei) are predominant saprobic on decaying mangroves and marine substrata, while only Aquilomyces patris is a root endophyte of white poplar (Fryar, Réblová & Catcheside, 2023).
The multigene phylogeny in the present study showed that our new fungus H. satunensis forms a distinct lineage within the genus Helicascus with robust statistical support (100% ML bootstrap and 1.00 Bayesian posterior probability). The DNA sequences of H. satunensis differ from those of H. kanaloanus and other species in terms of the number of nucleotide base substitutions in all the DNA regions, which indicates that these species are different. In conclusion, with its unique morphological and multigene phylogeny, we introduce H. satunensis as a novel mangrove fungus.