Identification, classification, and expression profile analysis of heat shock transcription factor gene family in Salvia miltiorrhiza
- Published
- Accepted
- Received
- Academic Editor
- Sushma Naithani
- Subject Areas
- Bioinformatics, Genomics, Molecular Biology, Plant Science
- Keywords
- Salvia miltiorrhiza, SmHSF, Gene expression, Drought, Heat
- Copyright
- © 2022 Liu et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2022. Identification, classification, and expression profile analysis of heat shock transcription factor gene family in Salvia miltiorrhiza. PeerJ 10:e14464 https://doi.org/10.7717/peerj.14464
Abstract
In response to abiotic stresses, transcription factors are essential. Heat shock transcription factors (HSFs), which control gene expression, serve as essential regulators of plant growth, development, and stress response. As a model medicinal plant, Salvia miltiorrhiza is a crucial component in the treatment of cardiovascular illnesses. But throughout its growth cycle, S.miltiorrhiza is exposed to a series of abiotic challenges, including heat and drought. In this study, 35 HSF genes were identified based on genome sequencing of Salvia miltiorrhiza utilizing bioinformatics techniques. Additionally, 35 genes were classified into three groups by phylogeny and gene structural analysis, comprising 22 HSFA, 11 HSFB, and two HSFC. The distribution and sequence analysis of motif showed that SmHSFs were relatively conservative. In SmHSF genes, analysis of the promoter region revealed the presence of many cis-acting elements linked to stress, hormones, and growth and development, suggesting that these factors have regulatory roles. The majority of SmHSFs were expressed in response to heat and drought stress, according to combined transcriptome and real-time quantitative PCR (qRT-PCR) analyses. In conclusion, this study looked at the SmHSF gene family using genome-wide identification, evolutionary analysis, sequence characterization, and expression analysis. This research serves as a foundation for further investigations into the role of HSF genes and their molecular mechanisms in plant stress responses.
Introduction
Plants are vulnerable to various stresses as a result of global warming, such as drought and heat (Lesk, Rowhani & Ramankutty, 2016; Yao et al., 2021). The intrinsic stress resistance systems allow plants to adapt continually in the face of various challenges, creating a complex web of stress responses (Chen & Zhu, 2004). Moreover, the crucial loop at the heart of this response network is transcription factors. In addition to these vast families of NAC, AP2/ERF, WRKY, and MYB (Ambawat et al., 2013; Jiang et al., 2017; Mizoi, Shinozaki & Yamaguchi-Shinozaki, 2012; Nakashima et al., 2012), a family of transcription factors called heat shock factors (HSFs) is known to have a role in plants’ response to stimuli (Guo et al., 2016). HSFs are part of intricate signaling networks that regulate reactions to a range of abiotic stresses, including cold, high temperatures, drought, hypoxia, and soil salinity (Andrási, Pettkó-Szandtner & Szabados, 2021).
Previous research has demonstrated that the HSF proteins in plants are made up of five motifs, the most conserved of which is an N-terminal DNA binding domain (DBD) (Sakurai & Enoki, 2010; Scharf et al., 2012), which is connected by a flexible linker to a bipartite heptad mode oligomerization domain (OD) with hydrophobic amino acid residues (HR-A/B region) (Baniwal et al., 2004). The other three are a nuclear localization signal (NLS), a nuclear export signal (NES), and an activation peptide motif (AHA) (Baniwal et al., 2004; Döring et al., 2000; Guo et al., 2016). In accordance with the number of amino acids inserted into the HR-A/B region and the length of the flexible linker region between the DBD and the HR-A/B region, plant HSFs are divided into three classes: hSFA, HSFB, and HSFC (Kotak et al., 2004). Although the HR-A/B region of HSFB is small and identical to that of all non-plant HSFs, members of HSFA and class HSFC insert 21 (HSFAs) and 7 (HSFCs) with an extended HR-A/B region that contains amino acid residues situated between the HR-A and HR-B groups, respectively (Nover et al., 1996; Scharf et al., 2012). Notably, AHA motifs, which stimulate the transcription of heat shock proteins (HSPs) by binding to transcription protein complexes, are specifically found in class A members (Scharf et al., 2012).
The first HSF gene of the plant was cloned from tomato (Scharf et al., 1990). The HSF family of many more plants has been thoroughly investigated as genome sequencing efforts have progressed, including the model plant Arabidopsis, the crops rice and maize, as well as the fruits pear and pineapple (Busch, Wunderlich & Schöffl, 2005; Chauhan et al., 2011; Lin et al., 2011; Qiao et al., 2015; Wang et al., 2021). Heat stress controls most plant HSFs. After being exposed to heat stress, it was discovered that the transcript levels of the A2 and A6 HSFs were predominant in wheat. During drought and salt stress, numerous TaHSFA members, as well as B1, C1, and C2 members were also upregulated (Xue et al., 2014). Furthermore, it has been demonstrated that a variety of additional abiotic stressors, including heat, salt, and drought, as well as phytohormones including jasmonic acid (Ja), abscisic acid (ABA), salicylic acid (SA), and ethylene (ET), regulate plant HSF genes (Hu et al., 2015). Sesame has been shown to contain 30 genes that encode HSF domains. 90% of HSFs were identified to respond to drought stress by time course expression profiling. After working with several HSFA genes, it was inferred that class B-HSFs might be a significant regulator of drought response in Sesame (Dossa, Diouf & Cissé, 2016). Recently, 30 HSF transcription factors (PvHSF 1-30) were discovered in Phaseolus vulgaris. Co linearity analysis revealed that PvHSFs were involved in controlling the response to abiotic stresses, with the majority of PvHSFs differentially expressed under cold, heat, salt, and heavy metal stresses, indicating that PvHSFs may have various roles depending on the type of abiotic stress (Zhang et al., 2022).
Salvia miltiorrhiza (Sm), a model medical plant whose dried roots and rhizomes as medicinal parts are known as Danshen (Lu et al., 2020), is widely used in Asia to treat cardiovascular disease (Li, Xu & Liu, 2018), and Alzheimer’s disease (Zhang et al., 2016). Water-soluble phenolic acids and lipid-soluble diterpenoid tanshinones are the distinct secondary metabolites, and two classes into which the principal active ingredients of S.miltiorrhiza can be subdivided (Jiang, Gao & Huang, 2019). The diterpenoid compound tanshinones, which comprises tanshinone I, tanshinone II, cryptotanshinone, and dihydrotanshinone, among others, has been isolated in more than 40 various forms from S. miltiorrhiza (Ma et al., 2015). It has considerable anti-inflammatory, antitumor, and anti-antioxidant properties and has also been shown to have strong anticancer properties both in vitro and in vivo (Chen et al., 2014; Wang et al., 2020). Caffeic acid, salvianolic acids, rosmarinic acid (RA), and lithospermic acids are examples of substances that exhibit phenolic acid properties (Ma et al., 2015). These compounds have antioxidant, anticoagulant, and cell protective properties (Wang, Morris-Natschke & Lee, 2007). As the climate changes, the yield of Salvia miltiorrhiza is threatened by several concurrent stresses, including drought and high temperatures. To further produce S.miltiorrhiza cultivars with greater heat and drought resistance, it is crucial to understand how S.miltiorrhiza tolerates heat and drought.
Although it has been demonstrated that members of the HSF family respond to heat and drought stress, it is still unclear how they express themselves in response to heat stress and what molecular mechanisms drought stress in Salvia miltiorrhiza involves. In light of this, we discovered HSF family members from Salvia miltiorrhiza’s genome data and examined the expression of HSF in S.miltiorrhiza under drought stress based on transcriptome data. After that, Salvia miltiorrhiza seedlings were treated by heat stress and observed 35 SmHSF members’ expression levels. Additionally, we combined a bioinformatic study of SmHSFs, including an investigation of the gene structure, motif, phylogeny, and promoter. So, in addition to analyzing the HSF family in Salvia miltiorrhiza, this work also assesses the expression profile of this family under heat stress and drought stress, which offers a foundation and concepts for further research into the functions of specific members.
Materials & Methods
Identification of SmHSF members
Genome Warehouse, BIG Data Center, project number PRJCA003150, which is available at https://ngdc.cncb.ac.cn/bioproject/browse/PRJCA003150, provided the assembly and annotation data for Salvia miltiorrhiza Bunge (Lamiaceae) (Song et al., 2020). The S.miltiorrhiza genome data was screened for HSF genes using the hmmsearch function in HMMER 3.0 (Finn, Clements & Eddy, 2011), and HSF members were identified using the hidden Markov model (HMM) corresponding to the HSF domain (PF00447) that was downloaded from the PFAM database (https://pfam.xfam.org/). E value is under 0.005 and default parameters were used. The S.miltiorrhiza genome database was searched using the BLASTP tool to find similar sequences using the Arabidosis HSF amino acid sequences that were obtained from TAIR (http://www.arabidopsis.org). Additionally, the SMART (Letunic & Bork, 2018) and CDD programs (https://www.ncbi.nlm.nih.gov/cdd/) were used to detect DBD domains in all acquired SmHSF proteins. Using TBtools (Chen et al., 2020) and the physical locational data from the S.miltiorrhiza genome, all of the SmHSFs were mapped to the eight chromosomes and three scaffolds of S.miltiorrhiza (Zhang et al., 2021). In addition, the ExPasy (https://web.expasy.org/compute_pi/) program was used to explore the physical characteristics of predicted HSF proteins. Subcellular localization predictions were generated using Plant-mPLoc with default parameters (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/) (Chou & Shen, 2010).
Structural, motif, and phylogenetic analysis of SmHSF genes
By comparing predicted coding sequences, the exon-intron distribution of each SmHSF gene was drawn using TBtools (Chen et al., 2020). The online tool MEME5.4.1 (https://meme-suite.org/meme/tools/meme) was used to investigate the conserved motifs of the SmHSF protein sequences. The number of motifs being requested is 10, and the range for motif width is 6 to 50 (inclusive). The results were visualized using TBtools. Using ClustalW in MEGAX with default parameters (https://www.megasoftware.net/), multiple sequence alignments of HSF proteins from Arabidopsis thaliana, Oryza sativa (Chauhan et al., 2011), Solanum lycopersicum (Jin et al., 2020), and S. miltiorrhiza were carried out. The sequences used for alignment are listed in Table S1. The alignment results were used to construct a phylogenetic tree using the Maximum Likelihood method with 1000 bootstrap replicates. Additionally, the evolutionary tree was embellished using Evolview (https://www.evolgenius.info/evolview/) (Subramanian et al., 2019).
Cis-acting elements analysis of SmHSFs
The S.miltiorrhiza genome database provided the 2,000-bp sequence upstream and 200-bp sequence downstream from the transcription start site of each SmHSF gene. These sequences were used to find cis-acting regulatory elements with the online tool PlantCARE (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/). A number of significant cis-acting elements were counted for each SmHSF using TBtools (Chen et al., 2020).
Expression profiles of SmHSFs under drought stress based on transcriptome data
The transcriptomic data generated from the leaf and root of S. miltiorrhiza under moderate drought stress have been described previously (Li, 2020). TBtools was used to create a heat map, and after the data Log2(FPKM+1), row normalization was carried out, to make it easier to compare the expression trends of each SmHSF in leaves and roots and after drought stress.
Plant materials and heat treatments
For heat stress treatment trials, tissue culture seedlings with the same or equivalent growth vigor were chosen. The S.miltiorrhiza tissue culture seedlings, which were grown for two months on MS medium, were moved to an artificial climate chamber with a constant temperature of 42 °C and exposed to heat stress continuously for 24 h, and six-time points (0 h, 1 h, 2 h, 6 h, 12 h, and 24 h) were selected for sample collection. All samples had three biological duplicates and were immediately frozen in liquid nitrogen following collection, and then stored at −80 °C for RNA extraction.
RNA isolation, and cDNA synthesis
Total RNA from S. miltiorrhiza samples were isolated using the Plant Total RNA Isolation Kit Plus following the manufacturer’s protocol (Foregene, Ltd., Chengdu, China). cDNA synthesis was done by using 1 ug of the total RNA samples with RT Easy™ II Kit (Foregene, Ltd., Chengdu, China).
Quantitative Real-Time PCR (qRT-PCR)
Reverse transcribed cDNA products were used as templates of quantitative Real-Time PCR (qRT-PCR). The reaction was carried out on a CFX Opus Real-time PCR system using the Real-Time PCR Easy ™ -SYBR Green I (Foregene, Ltd., Chengdu, China) following the manufacturer’s instructions. NCBI-BLAST Primer designed the primers for 35 SmHSF genes. S. miltiorrhiza Actin was used as an endogenous control for the normalization of expression levels of genes (Jiang et al., 2020). The relative expression levels were calculated using the 2− ΔΔ Ct method. Data were analyzed using one-way ANOVA in GraphPad Prism 9 software (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001). To ensure reproducibility and dependability, three biological replications and three technical replications were implemented for each sample. The primers for the SmHSFs used for qRT-PCR analyses are listed in Table S2.
Results
Identification and chromosomal localization of SmHSF gene family
A total of 35 SmHSF members were found in the Salvia miltiorrhiza genome after blastp and hmmsearch. These genes include HSF family domains and have been validated by NCBI-CD search and SMART. They were given the names SmHSF1 to SmHSF35 based on the gene IDs found in the newly sequenced genome. The members of the SmHSF gene family are listed in Table S3 along with their coding DNA and protein IDs and sequences.
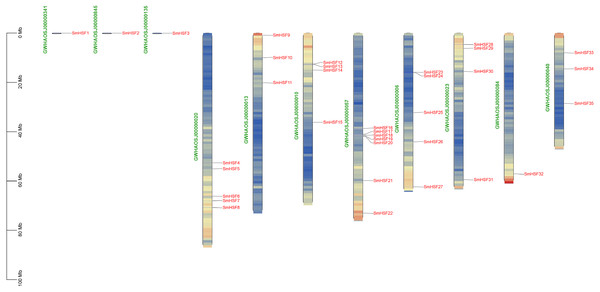
The 35 SmHSFs protein sequences ranged in length from 185 bp to 508 bp, with SmHSF11 having the longest and SmHSF24 having the shortest. SmHSF24 had a molecular weight of 21,538.39 Da, whereas SmHSF11 had a molecular weight of 56,360.16 Da. The highest and lowest members were consistent with the protein sequence. And the range of the isoelectric points (pI) was 4.64 (SmHSF25) to 9.93. (SmHSF29). All SmHSFs were determined to be unstable, with the exception of SmHSF35, which was stable, according to the examination of the instability index (the stability of the protein in a test tube). Additionally, the GRAVY ranged from −0.92 to −0.481, while the Aliphatic Index (AI) ranged from 57.61 to 78.99. Plant-mPLoc subcellular localization predictions suggested that all the HSF proteins were located in the nucleus. All of the information above is displayed in following Table 1. Except for SmHSF1, SmHSF2, and SmHSF3, which were found on the scaffolds, the remaining genes were determined to be spread among eight chromosomes according to the mapping of 35 SmHSFs on the Salvia miltiorrhiza chromosome (Fig. 1). Each chromosome contains a different number of S. miltiorrhiza HSF genes, and the position of the genes does not indicate anything about their function.
| Gene name | Protein length (aa) | Mw (Da) | pI | n.c.r | p.c.r | II | Stability | AI | GRAVY | Predicted location |
|---|---|---|---|---|---|---|---|---|---|---|
| SmHSF1 | 293 | 33682.07 | 8.43 | 47 | 49 | 44.92 | unstable | 57.61 | −0.872 | Nucleus |
| SmHSF2 | 332 | 38110.74 | 5.58 | 54 | 44 | 59.76 | unstable | 66.69 | −0.763 | Nucleus |
| SmHSF3 | 362 | 40936.75 | 4.84 | 60 | 41 | 55.29 | unstable | 70.25 | −0.665 | Nucleus |
| SmHSF4 | 290 | 31359.95 | 5.89 | 38 | 36 | 64.19 | unstable | 65 | −0.611 | Nucleus |
| SmHSF5 | 364 | 42052.88 | 5.84 | 62 | 55 | 55.31 | unstable | 76.59 | −0.707 | Nucleus |
| SmHSF6 | 341 | 37900.71 | 8.22 | 31 | 33 | 57.96 | unstable | 67.8 | −0.496 | Nucleus |
| SmHSF7 | 357 | 41644.68 | 5.72 | 60 | 52 | 61.34 | unstable | 61.23 | −0.92 | Nucleus |
| SmHSF8 | 233 | 26510.93 | 8.74 | 35 | 38 | 54.08 | unstable | 65.32 | −0.803 | Nucleus |
| SmHSF9 | 316 | 34592.59 | 4.88 | 54 | 38 | 52.21 | unstable | 78.99 | −0.481 | Nucleus |
| SmHSF10 | 345 | 37892.18 | 5.36 | 45 | 39 | 58.31 | unstable | 66.75 | −0.594 | Nucleus |
| SmHSF11 | 508 | 56360.16 | 5.26 | 67 | 49 | 61.03 | unstable | 76.38 | −0.559 | Nucleus |
| SmHSF12 | 328 | 37620.4 | 6.29 | 49 | 46 | 50.25 | unstable | 74.27 | −0.688 | Nucleus |
| SmHSF13 | 259 | 29742.92 | 9.19 | 37 | 42 | 52.13 | unstable | 77.53 | −0.725 | Nucleus |
| SmHSF14 | 482 | 53637.43 | 5.36 | 72 | 55 | 59.59 | unstable | 66.78 | −0.754 | Nucleus |
| SmHSF15 | 352 | 39746.64 | 5.92 | 46 | 38 | 56.96 | unstable | 71.7 | −0.628 | Nucleus |
| SmHSF16 | 388 | 43871.84 | 5.72 | 52 | 41 | 58.61 | unstable | 68.09 | −0.696 | Nucleus |
| SmHSF17 | 265 | 30541.47 | 6.72 | 34 | 33 | 53.25 | unstable | 71.4 | −0.685 | Nucleus |
| SmHSF18 | 252 | 28699.33 | 5.9 | 33 | 29 | 45.44 | unstable | 74.33 | −0.581 | Nucleus |
| SmHSF19 | 260 | 30162.06 | 5.77 | 35 | 32 | 48.55 | unstable | 78 | −0.662 | Nucleus |
| SmHSF20 | 187 | 21664.05 | 9.76 | 19 | 31 | 43.54 | unstable | 64.12 | −0.591 | Nucleus |
| SmHSF21 | 290 | 33416.89 | 6.92 | 33 | 32 | 63.92 | unstable | 71.24 | −0.638 | Nucleus |
| SmHSF22 | 425 | 48238.63 | 5.44 | 62 | 46 | 60.87 | unstable | 68.35 | −0.752 | Nucleus |
| SmHSF23 | 502 | 55086.51 | 4.78 | 68 | 44 | 61.74 | unstable | 62.01 | −0.627 | Nucleus |
| SmHSF24 | 185 | 21538.39 | 8.97 | 22 | 25 | 48.8 | unstable | 68.54 | −0.698 | Nucleus |
| SmHSF25 | 383 | 43547.77 | 4.64 | 65 | 39 | 47.29 | unstable | 71.31 | −0.653 | Nucleus |
| SmHSF26 | 227 | 26433.93 | 6.97 | 34 | 34 | 51.7 | unstable | 68.77 | −0.735 | Nucleus |
| SmHSF27 | 313 | 36130.43 | 5.11 | 55 | 39 | 44.08 | unstable | 66.68 | −0.792 | Nucleus |
| SmHSF28 | 399 | 45225.38 | 5.16 | 59 | 41 | 47.92 | unstable | 68.67 | −0.738 | Nucleus |
| SmHSF29 | 187 | 22125.45 | 9.93 | 21 | 35 | 62.15 | unstable | 65.24 | −0.9 | Nucleus |
| SmHSF30 | 336 | 37664.02 | 4.89 | 47 | 32 | 58.12 | unstable | 66.76 | −0.505 | Nucleus |
| SmHSF31 | 353 | 39554.09 | 5.7 | 48 | 39 | 59.9 | unstable | 64.67 | −0.724 | Nucleus |
| SmHSF32 | 487 | 54256.95 | 5 | 68 | 47 | 60.98 | unstable | 66.45 | −0.575 | Nucleus |
| SmHSF33 | 341 | 39147.22 | 5.39 | 64 | 49 | 52.12 | unstable | 62.02 | −0.734 | Nucleus |
| SmHSF34 | 224 | 25709.09 | 8.32 | 32 | 34 | 51.65 | unstable | 78.75 | −0.632 | Nucleus |
| SmHSF35 | 269 | 29506 | 6.34 | 35 | 34 | 34.25 | stable | 60.26 | −0.686 | Nucleus |
Notes:
- MW(Da)
-
Molecular weight in Dalton
- pI
-
isoelectric point
- n.c.r
-
total number of negatively charged residues (Asp + Glu)
- p.c.r
-
total number of positively charged residues (Arg + Lys)
- II
-
the instability index
- AI
-
Aliphatic index
- GRAVY
-
Grand average of hydropathicity
- Predicted location
-
Predicted Subcellar location
Figure 1: Distribution of SmHSF genes among 8 chromosomes and 3 scaffolds.
Phylogenetic, structural, and motif analysis of SmHSF genes
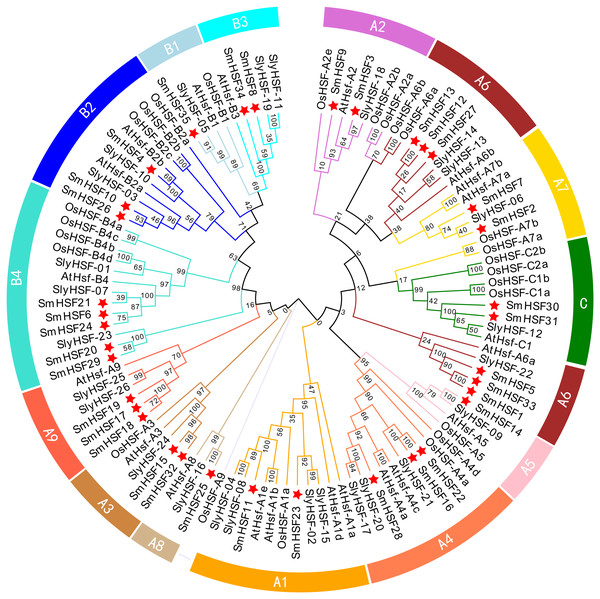
To investigate the phylogenetic relationship of 35 SmHSFs, a phylogenetic tree was constructed by combining SmHSFs with 21 Arabidopsis HSFs (AtHSFs), 25 rice HSFs (OsaHSFs), and 26 tomato HSFs (SlyHSFs) (Fig. 2). SmHSFs were separated into three groups (HSFA/HSFB/HSFC) in accordance with the grouping of HSFs in Arabidopsis. Each group has different member distributions. The distribution of members in these groups is like that in Arabidopsis thaliana, with Group A having the most members (22 genes) and Group C having the fewest members (two genes). Group A of SmHSF consisted of nine subgroups, from A1-A9; group B, with four subgroups (B1–B4); and group C, with only one subgroup (C1).
Figure 2: Phylogenetic relationships among the HSF family members of S. miltiorrhiza, Arabidopsis, rice, and tomato.
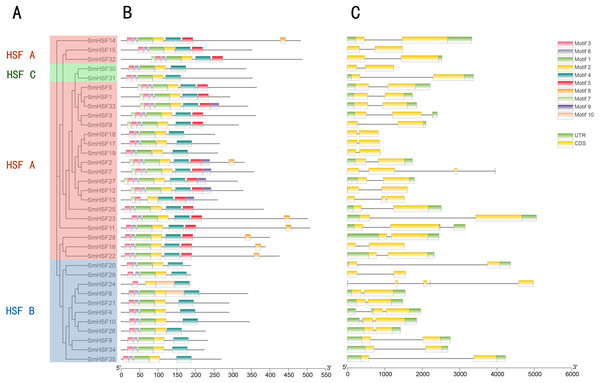
Full-length amino acid sequences were aligned using ClustalW and phylogenetic trees were constructed using the ML method in MEGAX. The tree clusters HSF proteins into different groups, which are represented by different colors within the branches.To further examine the structural diversity of genes, we analyzed the gene structures of 35 SmHSF genes. Analysis revealed that the majority of SmHSFs members belonged to the same subgroup and shared exon and intron structures (Fig. 3B). The number of exons and introns differs among SmHSFs, however. Of the 35 SmHSFs, 32 have two exons, two genes have three exons, and interestingly, SmHSF24 has five exon regions, two of which are incredibly short, and no UTR was discovered. All SmHSF genes have introns, of which 28 genes have one, five have two, one has three, and one has four.
Figure 3: Phylogenetic relationship, conserved protein motif structure and gene structure of SmHSFs.
(A) A phylogenetic tree was constructed based on the full-length sequence of Salvia miltiorrhiza HSFs protein using MEGA X software, including groups A, B, and C. (B) Motif composition of Salvia miltiorrhiza HSF protein. Motifs numbered 1–10 are shown in colored boxes, and the scale at the bottom indicates the length of the protein. Sequence information for each motif is in Table S4. (C) Gene structure of Salvia miltiorrhiza HSF gene.UTRs, untranslated regions, are represented by green boxes; CDS, coding sequences, are represented by yellow boxes. Black lines represent introns.Ten motifs were found in the SmHSF family members using motif analysis with MEME, the distribution of motifs corresponding to the phylogenetic tree of the SmHSFs family is shown in Fig. 3. Comparable motif compositions among the SmHSFs proteins clustered in the same subgroup imply that the members of the subgroup share similar activities. The DBD domain and HR-A/B are both present in the SmHSF family, just like the HSF family in other species. Motif2, motif3, and motif4 could be found in each member. Following sequence alignment, it was discovered that motif 1, motif 2, motif 3, and motif 6 make up the DBD domain of HSF, whereas motifs 4 and motifs 5 represent HR-A and HR-B, respectively. Except for SmHSF13 and SmHSF24, we detected motif1 and motif6 in all members. Only group A members possess motif 5, which has not been discovered in members of groups B or C. Additionally, only a few members of group A possess motifs 7, 8, and 9, where motif 8 is AHA and motif 9 is NLS. The sequence and distribution information of all motifs are in Table S4.
Cis-acting elements analysis of SmHSFs
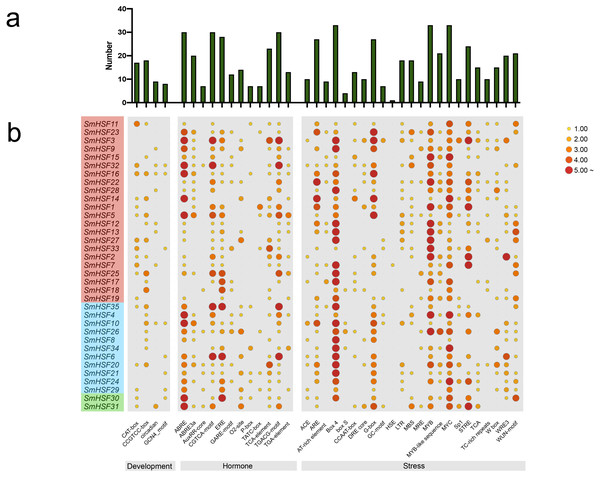
To further learn about the biological function of SmHSFs, the cis-acting elements in the 2,000 bp upstream and 200 bp downstream sequences from the transcription start sites of SmHSFs were examined using PlantCARE. A variety of cis-acting components, including those related to stress, hormones, and development, constitute the promoter of each SmHSF (Fig. 4). Members of SmHSFs are broadly diversified in terms of development-related components. The ABA-responsive element (ABRE), ABA and drought responsive element (ABRE3a), methyl jasmonate (MeJA)-responsive element (CGTCA-motif), and another MeJA-responsive element (TGACG motif), the ethylene-responsive element (ERE), gibberellin-responsive element (P-box), and cis-acting element involved in salicylic acid responsiveness (TCA-element) were all found in 30, 20, 30, 30, 28, 7, and 23 SmHSFs, respectively. Also, several light-associated cis-acting elements, like ACE and G-Box, as well as components connected to stress. MYB element, MYC element, DRE core (drought, salt, low temperature, and ABA responses), and STRE element (activated by heat shock, osmotic stress, low pH, and nutrient starvation) were identified in the promoters of 33, 33, 10, and 24 SmHSF genes, respectively. Additionally, anaerobic induction elements (AREs), low-temperature responsiveness elements (LTRs), MYB binding site involved in drought-inducibility (MBSs), defense and stress responsiveness (TC-rich repeats), wounding and pathogen responsiveness element (W-box), and wound-responsive element (WUN-motif) were found in 27, 18, 18, 10, 15, and 20 SmHSFs, respectively. However, the heat stress responsiveness element (HSEs) is only found in SmHSF7. These findings show that SmHSFs may be associated with a variety of transcriptional regulations involving hormones, stress responses, and development.
Figure 4: Analysis of the cis-acting elements in the promoter regions of the SmHSF genes.
(A) The graph was generated based on the presence of cis-acting element responsive to specific processes/elecitors/conditions (x-axis) in HSF gene family members (y-axis). (B) Heatmap of the number of cis-acting elements in each SmHSF. Based on the functional annotation, the cis-acting elements were classified into three major classes: development-, hormone-, stress-related cis-acting elements.Expression profiles of SmHSFs under drought stress
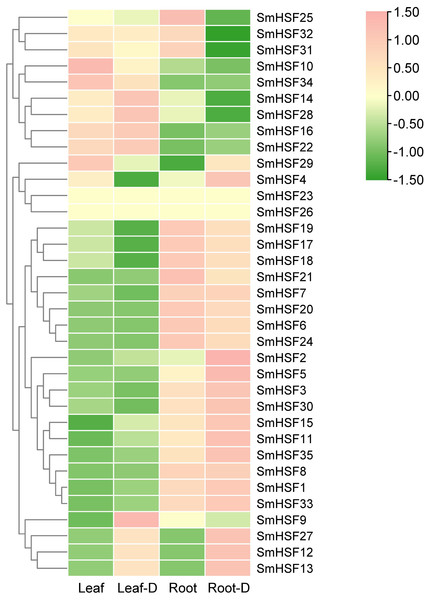
Tanshinone and salvianolic acid, have been observed to accumulate in the roots and leaves of S. miltiorrhiza, respectively (Sha, 2015; Wang & Wu, 2010). Additionally, drought stress will have some effect on HSF. To further investigate the expression of HSF in these two tissues and the expression changes in response to drought stress, we selected previous transcriptome data for HSF expression analysis. SmHSF23 and SmHSF 26 were not present in the transcriptome data, while other members were expressed to different degrees in roots and leaves as well as during drought stress. There were variations in the expression patterns of SmHSFs in roots and leaves, as shown in Fig. 5. SmHSF21 and SmHSF29 were not expressed in leaves or roots, respectively. There were 11 members, with the expression in roots being lower than in leaves. Of them, SmHSF10, SmHSF14, SmHSF16, and SmHSF34 clearly differed from the others. Notably, members like SmHSF20, SmHSF6, SmHSF8, SmHSF3, and SmHSF1 turned out to express mostly in the roots. Certain members like SmHSF10, SmHSF16, SmHSF30, SmHSF14, and SmHSF28 have varying expression in the roots and leaves and generally have greater expression levels. Following treatment for drought stress, it was found that the expression of 16 genes was up-regulated in the leaves, with SmHSF9 being the most visibly changed. It was discovered that the expression of 18 genes was up-regulated in roots, with SmHSF1, SmHSF2, SmHSF3, SmHSF5, and SmHSF33 standing out. Eleven SmHSF genes (SmHSF1, 2, 8, 11, 12, 13, 15, 16, 22, 27, and 35) are expressed in both roots and leaves under drought stress, and these genes may be activated by drought stress, further studies are required on how the functions.
Figure 5: Expression pattern of SmHSFs genes in Salvia miltiorrhiza leaf and root under drought stress.
Log2(FPKM+1) values were used for the heatmap. D, Drought. The color scale represents single row.Expression profiles of SmHSFs under heat stress
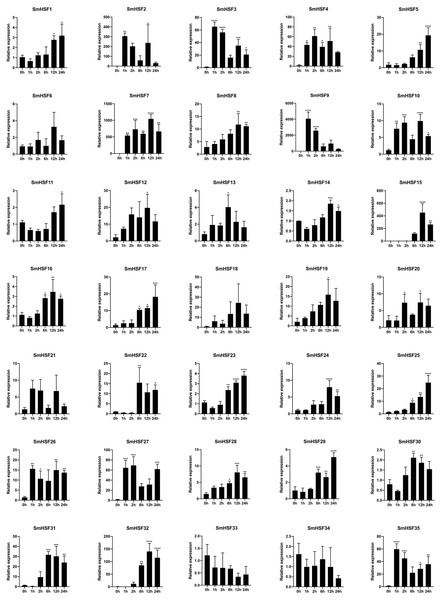
In response to heat stress, HSF genes are crucial for plant heat tolerance. To show how HSF genes react to heat stress in our work, the expression patterns of the SmHSFs gene family were identified using qRT-PCR. The results are depicted in Fig. 6. Despite barely being expressed, SmHSF25 rose in response to heat stress. When exposed to heat stress, the expression of SmHSF11, SmHSF14, and SmHSF30 did not change considerably, and the expression of SmHSF33, SmHSF34 were repressed, while the expression of other SmHSF genes was up-regulated to varying degrees. Notably, the expression of SmHSF2, SmHSF7, and SmHSF9 was markedly upregulated in response to heat stress, particularly SmHSF9, which was almost 4000-fold greater than the control at 2 h, showing that SmHSF2, SmHSF7, and SmHSF9 were implicated in the heat stress response pathway. In addition, the expression of SmHSF3, SmHSF4, SmHSF15, SmHSF27, SmHSF31, SmHSF32, and SmHSF35 also changed significantly after heat stress, and these genes are all worthy of further consideration.
Figure 6: Relative expression level of SmHSFs analyzed by qRT-PCR response to heat stress treatment.
qRT-PCR data were normalized using Salvia miltiorrhiza Actin gene and are shown relative to 0 h. X-axes are time course (0 h, 1 h, 2 h, 6 h, 12 h, and 24h) and y-axes are scales of relative expression level. All data represent means ±SD of three independent replicates.Statistical significance was analyzed by one-way ANOVA (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).Discussion
Transcription factors are crucial for plant growth, development, and stress resistance. Some transcription factor families in Salvia miltiorrhiza, including WRKY, AP2/ERF, MYB, bHLH, and NAC (Wu et al., 2021; Zhang et al., 2021) have been found, and yet nothing is known about the HSF transcription factor family. By controlling the expression of stress-responsive genes, HSFs play a crucial role in how plants react to various abiotic stimuli (Guo et al., 2016). In this study, we first discovered 35 HSF genes in Salvia miltiorrhiza and extensively analyzed these genes.
The 35 SmHSF genes are relatively small in number, being greater than those found in Arabidopsis (21), rice (25), and strawberry (17), but lower than those found in wheat (78) and Brassica juncea (60) (Hu et al., 2015; Li et al., 2020; Zhou et al., 2019). The S. miltiorrhiza HSF gene family was further separated into groups A, B, and C with molecular phylogenetic analysis. The largest group, group A, had 22 SmHSFs, which were separated into nine subgroups, A1 to A9. Group B, which contained 11 HSFs, including B1 to B4, came next. Only two individuals make up Group C, and all appear to be C1 members. Six individuals made up the Salvia miltiorrhiza A6 group, but it’s been previously demonstrated that members of this group are significantly less numerous in dicotyledonous plants than in monocotyledonous plants, indicating that this subgroup of Salvia miltiorrhiza may be the objective of more stringent purification selection (Wang et al., 2018). No members of the HSF-B5 and C2 subgroups were found in S. miltiorrhiza, which is following Arabidopsis, where C2 is a monocotyledon-specific gene that has only been detected in monocotyledonous plants (Guo et al., 2016; Wang et al., 2018). This uneven distribution of several groups demonstrates how their roles in the genome evolution of Salvia miltiorrhiza have changed over time.
By analyzing the structure and motif, the function of the gene was further defined. It was discovered that each subgroup had a similar distribution of the gene structure (Fig. 3), suggesting that the functions of the genes within the subgroup may be comparable. The DBD domain, which is highly conserved in plants, has about 100 amino acid residues (Scharf et al., 2012), as do the HSF genes in Salvia miltiorrhiza. On the other hand, the DBD domains of SmHSF13 and SmHSF24 only have 62 and 63 amino acid residues, respectively, indicating that their DBD domains are insufficient, which may be brought on by incomplete genome assembly. Only five members of group A had AHA motif identified, namely SmHSF11 from the A1 subgroup, SmHSF14 from the A5 subgroup, SmHSF16, SmHSF22, and SmHSF28 from the A4 subgroup, which all contribute to activator potential (Kotak et al., 2004). Without the AHA domain, other members may carry out their responsibilities by combining with other A-class HSFs to create hetero-oligomers, somewhat like B- and C-class HSFs (Guo et al., 2008a).
The analysis of the cis-acting elements in the promoter region identified numerous light responsive elements, MYB elements, MeJA elements, ABRE elements, STRE elements, and other elements, all of which suggest that these SmHSF genes are involved in hormone-related plant growth and development as well as in response to various stresses. The expression response of SmHSFs to heat and drought stress appears to be associated, particularly with those cis-acting components relevant to abiotic stress, such as MYB, MYC, ABRE, DRE, MBS, and STRE. The medicinal plant Salvia miltiorrhiza contains mostly water-soluble phenolic acids and fat-soluble tanshinones, which have been shown to accumulate predominantly in the roots and leaves. Previously, the HSF family was discovered to be tissue- and stage-specific in pineapple (Wang et al., 2021), Carnation (Li et al., 2019), and Tartary buckwheat (Liu et al., 2019). Although there were different expression levels in the roots and leaves of Salvia miltiorrhiza, it was discovered from the transcriptome data that the HSF gene did not appear to have any apparent tissue specificity, this finding was in line with that of soybean and sesame (Dossa, Diouf & Cissé, 2016; Lin et al., 2014). This indicated that the function of the HSF gene family was conserved in Salvia miltiorrhiza. 16 genes were discovered to be up-regulated in leaves, and 18 genes were found to be up-regulated in roots following treatment for drought stress; this up-regulation behavior was previously seen in Salix suchowensis (Zhang et al., 2015), and Chenopodium quinoa (Tashi et al., 2018). Numerous cis-acting elements linked to drought were found among the 11 SmHSFs whose expression was frequently increased following drought stress, including MYB, MYC, ABRE3a, DRE, and MBS, which were demonstrated to be linked to the regulation of HSF expression during drought (Li et al., 2019). These genes are expected to play a significant part in how S. miltiorrhiza responds to drought and other abiotic challenges (Zhu et al., 2017).
After heat treatment, 35 SmHSFs were investigated for expression. Among them, during heat stress treatment, 29 genes were up-regulated and two genes were down-regulated. SmHSF2, SmHSF7, and SmHSF9 are significantly up-regulated, particularly SmHSF9, which is almost 4000 times greater than the control at 2 h. The up-regulation of SmHSF7 also reached a peak at 12 h, which was about 1,000 times the relative expression of the control group, which may be related to the only HSE cis-acting element found in this gene, which significantly activated in response to heat stress. Along with HSE, STRE is a critical element of the heat stress response (Zhu et al., 2017). The Arabidopsis HSFA1a direct binding site for the STRE element was also discovered (Guo et al., 2008b). This suggests that STRE is present in those SmHSFs whose expression is up-regulated in response to heat stress, potentially pointing to differences in the regulation of several genes. Other genes whose expression value increased more than dozens of times after heat stress, such as SmHSF3, SmHSF4, SmHSF15, SmHSF27, SmHSF31, SmHSF32, and SmHSF35 are considered to be extremely sensitive to heat stress and play an essential role in response to heat stress. In fact, plants frequently experience many stresses along their development. SmHSF2, SmHSF8, SmHSF15, SmHSF27, SmHSF35, and other genes that respond to both heat and drought stress should be the main targets, and it is worthy of further research the regulatory mechanism of S.miltiorrhiza in response to abiotic stress.
Conclusions
Thirty-five SmHSF genes were identified from the Salvia miltiorrhiza genome, which were systematically examined using approaches such as chromosomal position, gene structure, motif, phylogenetic relationship, and cis-acting element analysis. RNA-seq data was used to examine the expression profile of the HSF gene in the roots and leaves of Salvia miltiorrhiza under drought stress. The changes in HSF under stress were evaluated in conjunction with the expression study of the HSF gene in S.miltiorrhiza under heat stress, which helped identify some methods that could increase Salvia miltiorrhiza tolerance to stress. Differences in the regulation of various genes may be explained by integrating the expression data with the findings of promoter analysis, which may also contribute to further screening of genes that are tolerant to heat and drought. The findings provide a basis for further studies on the function and molecular mechanism of HSF genes in plant stress response.
Supplemental Information
Protein sequences of other selected plant HSF genes used for phylogenetic analysis
List of the identified SmHSFs
The ID number and position of SmHSFs in the Salvia miltiorrhiza genome, and the protein and CDS sequences of SmHSFs.
Sequence and distribution information of all motifs in SmHSFs
Including the motif distribution of the SmHSF genes and 10 motif sequences analyzed by MEME
The promoter cis-acting elements of SmHSFs
Statistics on the number of cis-acting elements in SmHSFs promoters