Molecular phylogeny of Columbellidae (Gastropoda: Neogastropoda)
- Published
- Accepted
- Received
- Academic Editor
- Tim Collins
- Subject Areas
- Biodiversity, Evolutionary Studies, Marine Biology, Taxonomy, Zoology
- Keywords
- Marine Mollusca, Shell morphology, Gastropod classification, Radula
- Copyright
- © 2022 deMaintenon and Strong
- Licence
- This is an open access article distributed under the terms of the Open Government License.
- Cite this article
- 2022. Molecular phylogeny of Columbellidae (Gastropoda: Neogastropoda) PeerJ 10:e13996 https://doi.org/10.7717/peerj.13996
Abstract
The neogastropod family Columbellidae is a highly successful group of small, primarily epibenthic marine snails distributed worldwide and most abundant in the tropics. The great diversity of the group makes them attractive for studying evolutionary shifts in gastropod anatomy, morphology, ecology and diversity. The existing classification of the family has been based to a large degree on the morphology of the shell and radula. Indeed, membership in the family is traditionally confirmed using the unique morphology of the radula. To reconstruct columbellid phylogeny and assess monophyly of the group, we assembled a multilocus dataset including five mitochondrial and nuclear genes, for 70 species in 31 genera. Phylogenetic analyses using Bayesian inference and maximum likelihood are not well enough resolved to support a subfamilial classification, but do support the monophyly of the family and of several well-defined genera and supra-generic groupings. Two of the most diverse nominal genera, Mitrella and Anachis, are supported as highly polyphyletic. Overall, the resulting topologies indicate that the generic and subfamilial classification is in need of extensive revision but that phylogenomic data are needed to resolve columbellid relationships.
Introduction
Columbellids are a highly diverse neogastropod family of Buccinoidea, with 919 valid Recent species in 75 genera (MolluscaBase, 2021), distributed globally and represented in most marine habitats. The group is comprised of primarily small marine snails, some of which, being common and variable, are frequently used for shell craft. Columbellids are ecologically diverse and are mostly epibenthic on hard bottoms, but some are burrowers in soft sediments. Many occur in shallow water, with a substantial, still poorly documented fauna offshore to depths of 500 to 1,000 m (see e.g., Monsecour & Monsecour, 2016), and a few reach depths of 2,000 m (Bouchet & Warén, 1985). Most species are probably opportunistic carnivores, however some are herbivorous (Hatfield, 1979; Nielsen & Lethbridge, 1989; Kantor & Medinskaya, 1991).
Despite being diverse and sometimes very common where they occur, columbellid biology is generally poorly known and the relationships of columbellid taxa are essentially unresolved. This is most likely due to two factors. First, columbellids are too small to capture the attention of most collectors, with most species less than 15 mm in adult shell length. Second, they lack obvious conchological features that could be used to characterize the group, and instead vary greatly in shell form (see Figs. 1 and 2), tending to resemble species from many other caenogastropod families. In his taxonomic revision of Columbellidae, Radwin (1977a) listed 14 columbellid genera and six different caenogastropod families (Terebridae, Buccinidae, Olividae, Strombidae, Turridae and Conidae) with which they converge in shell form. To these we would add Mitridae, as columbellid specimens are often misplaced in this family as well and can only be differentiated by the lack of well-developed columellar plaits, though they often have weakly developed ones. Many columbellid species are easily confused with conoideans, as most possess posterior apertural canals very similar to members of that group, and there are columbellid genera with coniform (e.g., Parametaria Dall, 1916 and Conella Swainson, 1840) and terebriform (e.g., Mazatlania Dall, 1900) shells. An alleged Terebra species in the Mediterranean, upon further study (Bouchet & Gofas, 1983), was found to be a mislocalized record of the tropical American columbellid species Mazatlania cosentini (Philippi, 1836), which in fact was originally described as a Terebra.
The Columbellidae was originally established as a subfamily of Strombidae by William Swainson (1840) to contain a collection of species, in five genera, in which the shells are small, have the aperture edge internally thickened or striated, inflexed and denticulate, the inner lip ‘doubly toothed’, and a very small operculum. Two of the originally included genera (Crassispira and Pusiostoma, both of Swainson, 1840) have since been removed to other families (Pseudomelatomidae and Pisaniidae respectively). For a number of years, the family increased in content primarily through monographic works (e.g., Sowerby I, 1832; Sowerby I , 1844; Duclos, 1840; Duclos, 1846; Kiener , 1841; Gaskoin, 1852; Reeve, 1859). These works placed most species now recognized as columbellids in the catch-all genus Columbella Lamarck, 1799.
Meanwhile, attempts to classify the increasing number of species into genera were somewhat slower in coming. Adams & Adams (1853), based on shell characters, classified the group (∼240 species at the time) as a subfamily of Mitridae, with five genera and a number of subgenera. Carpenter (1856), in contrast, considered columbellids to belong to several different families, based primarily on shell and opercular characters. Tryon (1883) drew attention to the unique radular morphology typical of the group (but also included the buccinid genus Engina in Columbellidae based on its similar small size and shell morphology) and distributed the approximately 750 species into six subgenera of the genus Columbella, with twelve sections. Pace (1902) provided alphabetic lists of all living and fossil species and genus-group taxa (excluding Engina); he attempted to assess the validity of the included species but did not suggest a classification of the group.
Beginning in the late 1800s, the morphology of the radula gained attention as a taxonomically informative character set. The columbellid radula (see Fig. 3) is rachiglossan, like that of other buccinoids, but with a unique form. It consists of a rectangular, acuspate center plate and tall, sigmoid lateral teeth on a narrow base. The lateral teeth have one or two cusps, with the largest on the inside, and this larger cusp has two to four secondary cusps oriented orthogonally to the primary cusp axis. Mörch (1859) noted the unique character of the columbellid radula and used that to exclude several species currently placed in Buccinidae and other families. Troschel (1856–1879) described and illustrated the radular morphology in ten species and indicated that its basic form was key to establishing whether a species belongs in Columbellidae. Based on differences in radular morphology, he grouped the species of the family into two genera, Columbella and Pyrene Röding, 1798, each with two or more subgenera. Some Australian and New Zealand authors (e.g., Verco, 1910; Iredale, 1929) picked up on this usage and began to place most species in Pyrene, primarily because the type species of Pyrene (Pyrene punctata (Bruguière, 1789), Fig. 1E) is Indo-Pacific whereas that of Columbella (Columbella mercatoria (Linnaeus, 1758), Fig. 1A) is from the western Atlantic (Iredale, 1929). Suter (1909, 1913) also coined the new family group name Pyrenidae under the mistaken consideration that this was necessary by virtue of the name Pyrene being older than Columbella. However, most authors from both northern and southern hemispheres (e.g., Sowerby III, 1892; Pilsbry, 1895; Kobelt, 1892–1897; Hervier, 1899) continued to place most species in the family in a single genus. Lacking a generally accepted taxonomy, this was usually Columbella, however the usage of Pyrene also became common.
Figure 1: Shells of representative columbellids.
(A) Columbella mercatoria (Linnaeus, 1758), MNHN IM-2013-9415, Guadeloupe, 16.65 mm. (B) Nitidella nitida (Lamarck, 1822), MNHN IM-2013-8356 (juvenile), Malendure, Guadeloupe, 10.83 mm. (C) Euplica turturina (Lamarck, 1822), UF 410894B, Kiritimati, 9.76 mm. (D) Metanachis jaspidea (Sowerby I, 1844), MNHN IM-2013-13320, Papua New Guinea, 11.85 mm. (E) Pyrene punctata (Bruguière, 1789), UF 410842 (juvenile), Philippines, 12.45 mm. (F) Aesopus cumingii (Reeve, 1859), MNHN IM-2013-10364, Papua New Guinea, 13.95 mm. (G) Mitrella alofa (Hedley, 1899), MNHN IM-2013-16683, Papua New Guinea, 15.80 mm. (H) Pardalinops marmorata (Gray, 1839), MNHN IM-2013-1093, Papua New Guinea, 12.20 mm. (I) Graphicomassa ligula (Duclos, 1840), MNHN IM-2013-12892, Papua New Guinea, 19.55 mm. (J) Mitrella mindorensis (Reeve, 1859), MNHN IM-2013-14876, Papua New Guinea, 7.70 mm. (K) Mitrella moleculina (Duclos, 1840), MNHN IM-2013-1731, Papua New Guinea, 6.58 mm. (L) Pseudanachis basedowi (Hedley, 1918), SBMNH 199996, Prince Frederick Harbor, Western Australia, 14.70 mm.Figure 2: Shells of representative columbellids.
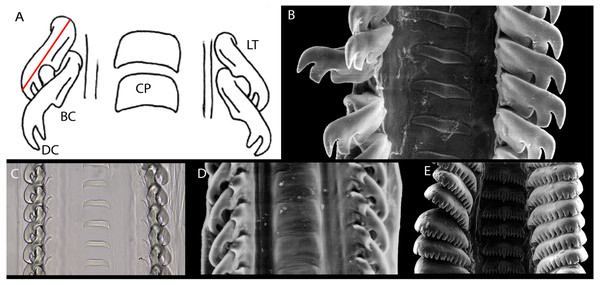
A: Anachis pardalis (Hinds, 1843), USNM 1231330, Panama, 8.57 mm; B: Costoanachis hotessieriana (d’Orbigny, 1842), MNHN IM-2013-20756, Guadeloupe, 6.33 mm; C. Decipifus sp., MNHN IM-2013-8302, Guadeloupe, 4.27 mm; D: Nassarina metabrunnea Dall & Simpson, 1901, MNHN IM-2013-20406, Guadeloupe, 6.95 mm; E. Steironepion monilifera (Sowerby I, 1844), MNHN IM-2019-1834, Guadeloupe, 4.87 mm; F. Suturoglypta pretrii (Duclos in Chenu, 1846), MNHN IM-2013-9260, Guadeloupe, 7.58 mm. G: Rhombinella laevigata (Linnaeus, 1758), MNHN IM-2013-20346 (juvenile), Guadeloupe, 13.25 mm; H: Zafrona isomella (Duclos, 1840), MNHN IM-2013-2315, Papua New Guinea, 4.13 mm; I: Falsuszafrona pulchella (Blainville, 1829), MNHN IM-2013-7890, Guadeloupe, 9.1 mm; J: Pyreneola cf. lozoueti Drivas & Jay, 1997, MNHN IM-2009-13250, Vanuatu, 4.43 mm; K: Seminella virginea (Gould, 1860), MNHN IM-2013-2357, Papua New Guinea, 3.23 mm; L: Pyreneola leptalea (E.A. Smith, 1902) MNHN IM-2019-1835, Mozambique, 2.58 mm.Figure 3: Columbellid radulae.
(A) Diagram of a generalized Mitrella radula. The red line indicates the axis of the primary lateral tooth cusp and its length. BC, basal secondary cusp; CP, center plate; DC, distal secondary cusp; LT: lateral tooth. (B) Columbella major, CAS 085593, LT length 189 µm. C. Anachis scalarina, USNM 1516834, LT length 75 µm. D., Amphissa columbiana, CAS 085592, LT length 91 µm. E., Pseudanachis basedowi, ANSP A-3390, LT width 130 µm.More recently, research on columbellid anatomy showed that columbellid reproductive anatomy varied significantly and might be informative for classification. The studies by Marcus & Marcus (1962, 1964) on nine columbellid species suggested two basic anatomical patterns in reproductive anatomy: species with a secondary spermatic vesicle and no prostatic gland in males, and an albumen gland and well developed bursa copulatrix in females (in species of Anachis, Costoanachis and Mitrella); versus those with a prostatic gland but no secondary spermatic vesicle in males, and females with no albumen gland or bursa copulatrix, with sperm storage and discharge occurring via the pericardium and pericardial-pallial duct (in species of Astyris, Columbella and Parvanachis). Research on additional species (Houston, 1976) however suggested that some of these structures are variably present within genera. An attempt to use characters from columbellid anatomy to generate a phylogeny for the family (deMaintenon, 1999), found that anatomical, radular, and conchological characters are all similarly homoplastic. Results of that study indicated that monophyly of the family was supported by a combination of radular and anatomical characters, but no conchological characters; conchological characters tended to be more informative at the generic level.
The first attempt at classification of columbellids above the genus level was Cossmann (1901), who recognized two subfamilies based primarily on the length of the siphonal canal of the shell. Species included in Columbellinae have a short or very short canal, whereas those placed in his new subfamily Atiliinae have a longer, straighter canal. Atilia is currently considered a synonym of the genus Anachis H. Adams & A. Adams (MolluscaBase, 2021), which is itself the basis for another little-used family name, Anachidae (Golikov & Starobogatov, 1975). The latter was established based on a combination of conchological and anatomical characters, but Golikov & Starobogatov (1975) did not indicate specifically what taxa were included, and it has not been commonly used. It is considered a synonym of Atiliinae by Bouchet et al. (2017) and of Columbellidae by Kantor et al. (2021).
The most recent subfamily classification was advanced by Radwin (1977a). Using radular and conchological characters, and given the two basic radular morphologies recognized by Troschel (1856–1879) and Thiele (1924) to diagnose the genera Columbella and Pyrene, Radwin divided the known species of western Atlantic Columbellidae into two subfamilies Columbellinae and Pyreninae. The former is a less diverse group based on the largely herbivorous genus Columbella, which has more or less strombiform shell and relatively large, strap-like, robust lateral teeth hypothetically adapted to scraping algae from hard surfaces (e.g., Fig. 3B). Even though neogastropods are typically carnivores, herbivory has been reported to occur in a number of columbellid taxa, e.g., Columbella mercatoria and C. rustica, Rhombinella laevigata (Marcus & Marcus, 1962; Bandel, 1974), and Euplica bidentata (Nielsen & Lethbridge, 1989). Pyreninae as Radwin construed it is a more diverse and much more heterogeneous group, with a variety of shell forms and smaller, narrower, more delicate lateral teeth ostensibly adapted to a carnivorous diet (e.g., radulae of Anachis scalarina and Amphissa columbiana, Figs. 3C, 3D). This latter type of radula, having narrow lateral teeth with a basal cusp separated from two distal cusps, is common in many columbellids. Marcus & Marcus (1962) suggested a relationship between diet and radular morphology, and additional study (deMaintenon, 1999) supported this, but the systematic implications of such a relationship have not been explored with independent evidence. Radwin’s (1977a) tentative phylogeny was based on “morphological similarities [of shell and radula] and sequence of appearance in time” (p. 404) and had Columbellinae as a lineage within a paraphyletic Pyreninae. Furthermore, his phylogeny was established only for western Atlantic species. Despite these limitations, his classification has been widely adopted.
Given the great diversity combined with anatomical and ecological variability of the Columbellidae, the group has great potential as a model group for studying evolution of form, function and biodiversity. However, until recently no attempt had been made to evaluate the monophyly and relationships of the family or its constituent taxa using molecular data. Kantor et al. (2021) have partially addressed this need through a molecular phylogenetic analysis of Buccinoidea, including seven species of Columbellidae in six genera. Their findings support a monophyletic Columbellidae, as sister (without support) to the monophyletic nassariid taxon Cylleninae. Overall the whole Nassariidae + Columbellidae complex, while being monophyletic, defies robust internal resolution. The objective then of the present study was to generate a molecular phylogeny of the Columbellidae, using a broad sampling of species from as many genera as possible, to assess the monophyly and relationships of the family and particularly of its constituent genera.
Our dataset was constructed using partial sequences from five mitochondrial and nuclear loci that were chosen based on their utility for other family-level molluscan phylogenetic studies: the mitochondrial cytochrome c oxidase I (COI), 16S rRNA, and 12S rRNA genes, and the nuclear Histone H3 and 28S rRNA (28S) genes. The resulting phylogenetic framework is used to assess columbellid monophyly and compared to previous classifications based on anatomical and morphological characters.
Material and Methods
We assembled a five-gene mitochondrial and nuclear dataset for 70 columbellid species in 31 genera, and included representative outgroups from the Nassariidae, as suggested by the results obtained by Kantor et al. (2021); trees were rooted with Melongenidae. A total of 98 terminals were included; sequences were newly generated for 71 of these, and the remainder downloaded from GenBank. See Table 1 for sources.
| Species | Voucher ID | COI | 12S | 16S | H3 | 28S | Location, Station, Depth | Source |
|---|---|---|---|---|---|---|---|---|
| Melongenidae | ||||||||
| Hemifusus colosseus (Lamarck, 1816) | LSGB 23301 | HQ834068 | HQ833890 | HQ833939 | HQ834161 | – | Zou, Li & Kong (2011) | |
| Melongena patula (Broderip & Sowerby, 1829) | MZUR BAU00794 | FM999172 | FM999093 | FM999124 | – | FM999148 | Oliverio & Modica (2010) | |
| Nassariidae | ||||||||
| Buccinanops deformis (King, 1832) | MNHN IM-2009-24004 | KY451220 | KY488927 | KY488730 | KY489294 | KY489125 | Galindo et al. (2016) | |
| Bullia diluta (Krauss, 1848) | MNHN IM-2009-22535 | KY451224 | KY88930 | KY488733 | KY489298 | – | Galindo et al. (2016) | |
| Cyllene owenii Gray in Griffith & Pidgeon, 1834 | MNHN IM-2009-23727 | KY451236 | KY488941 | – | KY489308 | KY489131 | Galindo et al. (2016) | |
| Cyllene parvula Bozzetti, 2014 | MNHN IM-2009-12765 | KY451237 | KY488942 | KY488742 | KY489309 | KY489132 | Galindo et al. (2016) | |
| Dorsanum miran (Bruguière, 1789) | MNHN IM-2013-52428 | KY451239 | KY488944 | KY488744 | KY489311 | KY489134 | Galindo et al. (2016) | |
| Engoniophos unicinctus (Say, 1826) | MNHN IM-2009-24414 | KY451413 | KY489122 | KY488923 | KY489375 | – | Galindo et al. (2016) | |
| Nassarius arcularia (Linnaeus, 1758) | MNHN IM-2007-31898 | KY451259 | KY488968 | KY488766 | KY489317 | KY489155 | Galindo et al. (2016) | |
| Nassarius glans (Linnaeus, 1758) | MCZ 378603 | KT754006 | – | KT753883 | KT754135 | KT753774 | Couto et al. (2016) | |
| Nassarius niger (Hombron & Jacquinot, 1848) | MNHN IM-2007-31730 | KY451241 | KY488946 | KY488746 | KY489313 | KY489136 | Galindo et al. (2016) | |
| Naytia granulosa (Lamarck, 1822) | MNHN IM-2009-23948 | KY451225 | KY488931 | KY488734 | KY489299 | KY489128 | Galindo et al. (2016) | |
| Oligohalinophila dorri (Wattebled, 1886) | MNHN IM-2009-20649 | KY773620 | KY706413 | KY706391 | KY706452 | KY706430 | Galindo et al. (2016) | |
| Phos senticosus (Linnaeus, 1758) | LSGB 2320901 | HQ834064 | HQ833884 | HQ833934 | HQ834155 | – | Zou, Li & Kong (2011) | |
| Phrontis antillarum (d’Orbigny, 1847) | MNHN IM-2009-24320 | KY451258 | KY488967 | KY488765 | KY489316 | KY489154 | Galindo et al. (2016) | |
| Phrontis pagoda (Reeve, 1844) | MZUR BAU00237 | FM999173 | FM999094 | FM999125 | Oliverio & Modica (2010) | |||
| Reticunassa paupera (Gould, 1850) | MNHN IM-2007-31778 | KY499730 | KY489057 | KY488856 | KY489349 | KY489232 | Galindo et al. (2016), 2017 | |
| Tomlinia frausseni Thach, 2014 | MNHN IM-2013-52188 | KY451417 | – | KY488926 | KY489378 | Galindo et al. (2016) | ||
| Tritia obsoleta (Say, 1822) | MNHN IM-2009-21755 | KY451244 | KY488949 | KY488748 | KY489315 | KY489139 | Galindo et al. (2016) | |
| Tritia reticulata (Linnaeus, 1758) | MCZ 378509 | KT 753983 | – | KT754113 | KT753750 | Couto et al. (2016) | ||
| Columbellidae | ||||||||
| Aesopus cumingii (Reeve, 1859) | MNHN IM-2013-10364 | OM674535 | – | OM730102 | OM687436 | OM773498 | Papua New Guinea, PB04, 30 m | |
| Alia carinata (Hinds, 1844) | MIB 627 | KX069366 | – | – | – | KX070267 | Castelin et al. (2016) | |
| Amphissa columbiana Dall, 1916 | MIB 1161 | KX069398 | – | – | – | KX070275 | Castelin et al. (2016) | |
| Anachis cf. chuni (Thiele, 1925) | MNHN IM-2009-7412 | – | – | OM730079 | OM687413 | OM773479 | Mozambique, CC3171, 771–776 m | |
| Anachis fluctuata (G. B. Sowerby I, 1832) | USNM 1516848 | – | – | OM730091 | OM687424 | OM773488 | Panama, intertidal | |
| Anachis pardalis (Hinds, 1843) | USNM 1516838 | OM674520 | OM751364 | OM730080 | OM687414 | OM773480 | Panama, intertidal | |
| Anachis rugosa (G. B. Sowerby I, 1832) | USNM 1516840 | OM674521 | – | OM730081 | OM687415 | OM773481 | Panama, intertidal | |
| Anachis scalarina (G. B. Sowerby I, 1832) | USNM 1516834 | – | – | OM730082 | OM687416 | – | Panama, intertidal | |
| Anachis varia (G. B. Sowerby I, 1832) | USNM 1516844 | OM674523 | OM751365 | OM730084 | OM687417 | – | Panama, intertidal | |
| Anachis sp. | MNHN IM-2009-11313 | OM674522 | – | OM730083 | – | OM773482 | Solomon Islands, CP2767, 416–425 m | |
| Columbella aureomexicana (Howard, 1963) | MCZ 378333 | KT753999 | – | – | KT754128 | KT753766 | Couto et al. (2016) | |
| Columbella major G. B. Sowerby I, 1832 | USNM 1516842 | OM674526 | OM751368 | OM730087 | OM687420 | OM773484 | Panama, PV05, intertidal | |
| Columbella mercatoria (Linnaeus, 1758) | MNHN IM-2013-9415 | OM674527 | OM751369 | OM730088 | OM687421 | OM773485 | Guadeloupe, GD29, 4 m | |
| Costoanachis hotessieriana (d’Orbigny, 1842) | MNHN IM-2013-20756 | OM674529 | OM751370 | OM730092 | OM687425 | OM773489 | Guadeloupe, GD11, 14 m | |
| Decipifus sp. | MNHN IM-2013-8302 | – | OM751371 | OM730093 | OM687426 | – | Guadeloupe, GM07, 1 m | |
| Euplica aff. deshayesii (Crosse, 1859) | MNHN IM-2013-1084 | – | OM751372 | OM730094 | OM687427 | OM773490 | Papua New Guinea, PB12, 7–15 m | |
| Euplica deshayesii (Crosse, 1859) | MNHN IM-2013-10290 | OM674530 | OM751373 | OM730095 | OM687428 | OM773491 | Papua New Guinea, PR01, 22 m | |
| Euplica ionida (Duclos, 1840) | MNHN IM-2007-13197 | OM674531 | – | OM730096 | OM687429 | OM773492 | Vanuatu, DB16, 32–40 m | |
| Euplica turturina (Lamarck, 1822) | UF 410894A | – | – | – | – | OM773493 | Line Islands, BLINX-197, 0–1 m | |
| Euplica turturina (Lamarck, 1822) | UF 410894B | OM674532 | OM751374 | OM730097 | OM687430 | – | Line Islands, BLINX-197, 0–1 m | |
| Falsuszafrona idalina (Duclos, 1840) | MNHN IM-2013-20300 | OM674565 | OM751412 | – | OM687469 | OM773532 | Guadeloupe, GM07, 1 m | |
| Falsuszafrona pseudopulchella Pelorce, 2020 | MNHN IM-2013-7731 | – | OM751411 | OM730144 | OM687468 | OM773531 | Guadeloupe, GB17, 13 m | |
| Falsuszafrona pulchella (Blainville, 1829) | MNHN IM-2013-7890 | – | OM751414 | – | OM687471 | OM773534 | Guadeloupe, GM10, 1 m | |
| Graphicomassa adiostina (Duclos, 1840) | MNHN IM-2013-306 | OM674533 | OM751375 | OM730098 | OM687431 | OM773494 | Papua New Guinea, PB01, 6-10 m | |
| Graphicomassa ligula (Duclos, 1840) | MNHN IM-2013-12892 | OM674534 | OM751376 | OM730099 | OM687432 | OM773495 | Papua New Guinea, PM19, 0–1 m | |
| Indomitrella conspersa (Gaskoin, 1852) | MNHN IM-2013-13605 | – | OM751377 | OM730100 | OM687433 | OM773496 | Papua New Guinea, PD28, 1–4 m | |
| Indomitrella schepmani (K. Monsecour & D. Monsecour, 2007) | MNHN IM-2013-1040 | – | OM751378 | OM730101 | OM687434 | OM773497 | Papua New Guinea, PS07, 13 m | |
| Indomitrella sp. | MNHN IM-2009-12917 | – | OM751379 | – | OM687435 | – | Madagascar, CP3203, 50–52 m | |
| Metanachis jaspidea (Sowerby I, 1844) | MNHN IM-2013-13320 | OM674536 | – | OM730103 | OM687437 | OM773499 | Papua New Guinea, PR48, unknown | |
| Metanachis laingensis Sleurs, 1985 | MNHN IM-2013-1749 | – | OM751380 | OM730104 | OM687438 | OM773500 | Papua New Guinea, PB10, 10 m | |
| Microcithara harpiformis (G. B. Sowerby I, 1832) | USNM 1516850 | – | – | OM730105 | OM687439 | – | Panama, intertidal | |
| Mitrella alofa (Hedley, 1899) | MNHN IM-2013-16683 | OM674537 | OM751381 | OM730106 | OM687440 | OM773501 | Papua New Guinea, PP13, 120 m | |
| Mitrella antares Costa & Souza, 2001 | MNHN IM-2013-9001 | OM674525 | OM751367 | OM730086 | OM687419 | OM773483 | Guadeloupe, GB31, 15 m | |
| Mitrella bicincta (Gould, 1860) | LSGB 23102 | HQ834055 | HQ833864 | HQ833925 | HQ834136 | – | Zou, Li & Kong (2011) | |
| Mitrella burchardi (Dunker, 1877) | LSGB 23408 | HQ834098 | – | HQ833970 | HQ834191 | – | Zou, Li & Kong (2011) | |
| Mitrella delannoyei Pelorce, 2013 | MNHN IM-2009-31242 | OM674524 | OM751366 | OM730085 | OM687418 | – | Guadeloupe, GD70, 100 m | |
| Mitrella loyaltyensis (Hervier, 1899) | MNHN IM-2009-13261 | OM674538 | OM751382 | OM730108 | – | OM773502 | Vanuatu, FB92, 2–4 m | |
| Mitrella mindorensis (Reeve, 1859) | MNHN IM-2013-14876 | OM674539 | OM751383 | OM730109 | OM687442 | OM773503 | Papua New Guinea, PD49, 2–5 m | |
| Mitrella moleculina (Duclos, 1840) | MNHN IM-2013-1731 | OM674540 | OM751384 | OM730110 | OM687443 | OM773504 | Papua New Guinea, PB08, 4–5 m | |
| Mitrella aff. moleculina (Duclos, 1840) | MNHN IM-2013-17975 | OM674541 | OM751385 | OM730111 | OM687444 | OM773505 | Papua New Guinea, PS42, 18–27 m | |
| Mitrella nycteis (Duclos, 1846) | MNHN IM-2019-18326 | OM674528 | – | OM730089 | OM687422 | OM773486 | Guadeloupe, GS01, 3 m | |
| Mitrella nympha (Kiener, 1841) | MNHN IM-2013-1875 | OM674542 | OM751386 | OM730112 | – | OM773506 | Papua New Guinea, PM12, 0–1 m | |
| Mitrella ocellata (Gmelin, 1791) | MNHN IM-2013-20488 | OM674543 | OM751387 | OM730113 | OM687445 | OM773507 | Guadeloupe, GM07, 1 m | |
| Mitrella ocellata (Gmelin, 1791) | USNM 1516846 | – | – | OM730114 | OM687446 | OM773508 | Florida, intertidal | |
| Mitrella scripta (Linnaeus, 1758) | MCZ 378586 | KT754022 | – | KT753895 | KT754151 | KT753791 | Couto et al. (2016) | |
| Mitrella sp. | MNHN IM-2009-11299 | – | – | OM730115 | OM687447 | OM773509 | Solomon Islands, CP2817, 1136–1750 m | |
| Mitrella sp. | MNHN IM-2009-12918 | – | – | OM730107 | OM687441 | – | Madagascar, CP3208, 231–237 m | |
| Nassarina metabrunnea Dall & Simpson, 1901 | MNHN IM-2013-20406 | – | OM751388 | OM730116 | OM687448 | OM773510 | Guadeloupe, GD01, 80 m | |
| Nitidella nitida (Lamarck, 1822) | MNHN IM-2013-8356 | – | OM751389 | OM730117 | OM687449 | OM773511 | Guadeloupe, GM07, 1 m | |
| Pardalinops marmorata (Gray, 1839) | MNHN IM-2013-1093 | – | OM751390 | OM730118 | OM687450 | OM773512 | Papua New Guinea, PB14, 15 m | |
| Pardalinops testudinaria (Link, 1807) | MNHN IM-2013-12945 | OM674544 | OM751391 | OM730119 | OM687451 | OM773513 | Papua New Guinea, PM19, 0–1 m | |
| Parvanachis obesa (C. B. Adams, 1845) | MNHN IM-2009-31220 | – | OM751392 | OM730121 | OM687453 | OM773514 | Guadeloupe, GD49, 3 m | |
| Parvanachis sp. | MNHN IM-2009-31185 | – | – | OM730120 | OM687452 | – | Guadeloupe, GM03, 1 m | |
| Pseudamycla formosa (Gaskoin, 1852) | LSGB 23409 | HQ834097 | HQ833920 | HQ833969 | HQ834190 | – | Zou, Li & Kong (2011) | |
| Pseudanachis basedowi (Hedley, 1918) | SBMNH 199995 | – | OM751393 | OM730122 | OM687454 | OM773515 | Western Australia, WP01, intertidal | |
| Pyrene flava (Bruguière, 1789) | MNHN IM-2009-12920 | OM674545 | – | OM730123 | OM687455 | OM773516 | Madagascar, DW3237, 50–107 m | |
| Pyrene obtusa (G. B. Sowerby I, 1832) | UF 400892 | OM674546 | OM751394 | OM730124 | – | OM773517 | Society Islands, BMOO-981, 0–1 m | |
| Pyrene punctata (Bruguière, 1789) | UF 410842 | OM674547 | OM751395 | OM730125 | OM687456 | OM773518 | Philippines, Bbol13, 5–10 m | |
| Pyrene cf. punctata (Bruguière, 1789) | MNHN IM-2013-11205 | OM674548 | OM751396 | OM730126 | OM687457 | OM773519 | Papua New Guinea, PR11, 40 m | |
| Pyreneola leptalea (E.A. Smith, 1902) | MNHN IM-2019-1835 | OM674562 | OM751408 | OM730141 | OM687466 | OM773529 | Mozambique, MM04, 0–1 m | |
| Pyreneola cf. lozoueti Drivas & Jay, 1997 | MNHN IM-2009-13250 | OM674549 | – | OM730127 | – | – | Vanuatu, EP35, 10–51 m | |
| Pyreneola melvilli (Hedley, 1899) | MNHN IM-2009-12974 | OM674550 | OM751397 | OM730128 | – | – | Vanuatu, NB12, 20 m | |
| Rhombinella laevigata (Linnaeus, 1758) | MNHN IM-2013-20346 | OM674551 | OM751398 | OM730129 | OM687458 | OM773520 | Guadeloupe, GM07, 1 m | |
| Seminella peasei (Martens & Langkavel, 1871) | USNM 1516836 | OM674552 | OM751399 | – | OM687459 | OM773521 | Hawaii, HR01, 2–3 m | |
| Seminella virginea (Gould, 1860) | MNHN IM-2013-2357 | OM674553 | OM751400 | OM730130 | – | OM773522 | Papua New Guinea, PB16, 5 m | |
| Steironepion moniliferum (Sowerby I, 1844) | MNHN IM-2019-1834 | – | OM751401 | OM730131 | OM687460 | OM773523 | Guadeloupe, GD63, 20 m | |
| Sulcomitrella aff. circumstriata (Schepman, 1911) | MNHN-IM-2009-11298 | OM674554 | – | – | – | – | Solomon Islands, CP2848, 414–456 m | |
| Sulcomitrella aff. circumstriata (Schepman, 1911) | MNHN IM-2009-11302 | – | OM751402 | OM730132 | – | OM773524 | Solomon Islands, CP2837, 381–422 m | |
| Sulcomitrella monodonta (Habe, 1958) | MNHN IM-2009-13294 | OM674556 | OM751403 | OM730134 | OM687462 | OM773526 | Philippines, CP2720, 300-301 m | |
| Sulcomitrella monodonta (Habe, 1958) | MNHN IM-2009-11304 | OM674557 | OM751404 | OM730135 | – | – | Solomon Islands, CP2282, 150–160 m | |
| Sulcomitrella cf. rosadoi (Bozzetti, 1998) | MNHN IM-2009-12924 | OM674555 | – | OM730133 | OM687461 | OM773525 | Madagascar, CP3289, 332–379 m | |
| Sulcomitrella sp. | MNHN IM-2009-7399 | – | – | OM730090 | OM687423 | OM773487 | Mozambique, DW3169, 450 m | |
| Suturoglypta pretrii (Duclos, 1846) | MNHN IM-2013-9260 | OM674558 | – | OM730136 | OM687463 | OM773527 | Guadeloupe, GD49, 3 m | |
| Zafra ambonensis deMaintenon, 2008 | MNHN IM-2007-13217 | – | OM751405 | OM730137 | – | – | Vanuatu, DB46, 2–3 m | |
| Zafra cf. hahajimana (Pilsbry, 1904) | MNHN IM-2009-13051 | OM674560 | OM751406 | OM730139 | OM687464 | OM773528 | Philippines, M10, 3 m | |
| Zafra hahajimana (Pilsbry, 1904) | MNHN IM-2009-13237 | OM674559 | – | OM730138 | – | – | Vanuatu, EP01, 46–47 m | |
| Zafra hervieri (Pace, 1903) | MNHN IM-2007-13222 | OM674561 | OM751407 | OM730140 | OM687465 | – | Vanuatu, DB46, 2–3 m | |
| Zafra ocellatula (Hervier, 1900) | MNHN IM-2013-6206 | OM674563 | OM751409 | OM730142 | OM687467 | – | Papua New Guinea, PB47, 5 m | |
| Zafra pumila (Dunker, 1858) | MNHN IM-2009-13047 | OM674564 | OM751410 | OM730143 | – | OM773530 | Philippines, M10, 3 m | |
| Zafrona isomella (Duclos, 1840) | MNHN IM-2013-2315 | OM674566 | OM751413 | OM730145 | OM687470 | OM773533 | Papua New Guinea, PR22, 3–10 m |
Notes:
- LSGB
-
Laboratory of Shellfish Genetics and Breeding, Fisheries College, Ocean University of China, Qingdao, China
- MCZ
-
Museum of Comparative Zoology, Harvard University
- MIB
-
Fisheries and Oceans Canada, Pacific Biological Station
- MNHN
-
Muséum National d’Histoire Naturelle, Paris
- MZUR
-
Zoological Museum of “La Sapienza” Roma University
- SBMNH
-
Santa Barbara Museum of Natural History
- UF
-
Florida Museum of Natural History, University of Florida
- USNM
-
National Museum of Natural History, Washington, DC
Taxon sampling
The specimens used in this study were obtained primarily from the collections of the Muséum National d’Histoire Naturelle in Paris (MNHN), supplemented with some from the Florida Museum of Natural History (UF) and Santa Barbara Museum of Natural History (SBMNH), and some material from the personal collections of MJdeM, which has been vouchered at the National Museum of Natural History in Washington, DC (USNM). Most of these materials were collected via biodiversity surveys in the tropics, so cold water taxa are not well represented. Extant Columbellidae have their greatest diversity in the tropics however, with almost 90% of named species being tropical or warm temperate (MJdeM, unpubl.). Higher latitudes tend to be dominated by members of specific genera, for example Amphissa (included in our analysis) and Astyris H. Adams & A. Adams (1853) mostly from the North Pacific and North Atlantic, and Liratilia Finlay, 1927 and Macrozafra Finlay, 1926 from temperate waters of Australia and New Zealand. The objective in selecting taxa was to sample: (1) as many representatives of type species of available genus-group names as possible (see Table 2), and, (2) at least two species from each nominal taxon in the genus group to test monophyly of those taxa.
| Genus | Type species |
|---|---|
| AesopusGould, 1860 | Aesopus japonicus Gould, 1860 |
| AliaH.Adams & A. Adams,1853 | Alia carinata (Hinds, 1844) |
| AmphissaH.Adams & A. Adams,1853 | Amphissa columbiana Dall, 1916 |
| AnachisH.Adams & A. Adams,1853 | Anachis scalarina (G. B. Sowerby I, 1832) |
| Atilia H. Adams & A. Adams,1853 | Columbella suffusa G. B. Sowerby I, 1844 (taxon inquirendum) |
| Colombellarius Duméril, 1805 | Columbella mercatoria (Linnaeus, 1758) |
| ColumbellaLamarck, 1799 | Columbella mercatoria (Linnaeus, 1758) |
| Conidea Swainson, 1840 | Pyrene punctata (Bruguière, 1789) |
| CostoanachisSacco, 1890 | Costoanachis saccostata Radwin, 1977 |
| DecipifusOlsson & McGinty, 1958 | Decipifus sixaolus Olsson & McGinty, 1958 |
| EuplicaDall, 1889 | Euplica turturina (Lamarck, 1822) |
| FalsuszafronaPelorce, 2020 | Falsuszafrona idalina (Duclos, 1840) |
| GraphicomassaIredale, 1929 | Graphicomassa ligula (Duclos, 1840) |
| IndomitrellaOostingh, 1940 | Indomitrella puella (Sowerby I, 1844) |
| MetanachisThiele, 1924 | Metanachis jaspidea (Sowerby I, 1844) |
| MicrocitharaP. Fischer, 1884 | Microcithara harpiformis (Sowerby I, 1832) |
| Mitrella Risso, 1826 | Mitrella scripta (Linnaeus, 1758) |
| NassarinaDall, 1889 | Nassarina bushiae (Dall, 1889) |
| NitidellaSwainson, 1840 | Nitidella nitida (Lamarck, 1822) |
| PardalinopsdeMaintenon, 2008 | Pardalinops testudinaria (Link, 1807) |
| ParvanachisRadwin, 1968 | Parvanachis obesa (C. B. Adams, 1845) |
| PseudamyclaPace, 1902 | Pseudamycla dermestoidea (Lamarck, 1822) |
| PseudanachisThiele, 1924 | Pseudanachis basedowi (Hedley, 1918) |
| PyreneRöding, 1798 | Pyrene punctata (Bruguière, 1789) |
| PyreneolaIredale, 1918 | Pyreneola abyssicola (Brazier, 1877) |
| RhombinellaRadwin, 1968 | Rhombinella laevigata (Linnaeus, 1758) |
| SeminellaPease, 1868 | Seminella peasei (Martens & Langkavel, 1871) |
| SteironepionPilsbry & Lowe, 1932 | Steironepion piperatum (E. A. Smith, 1882) |
| SulcomitrellaKuroda, Habe & Oyama, 1971 | Sulcomitrella monodonta (Habe, 1958) |
| SuturoglyptaRadwin, 1968 | Suturoglypta pretrii (Duclos, 1846) |
| ZafraA. Adams, 1860 | Zafra mitriformis A. Adams, 1860 |
| ZafronaIredale, 1916 | Zafrona isomella (Duclos, 1840) |
DNA sequencing
Protocols for DNA extraction and sequencing at the Laboratories of Analytical Biology at the USNM followed those in Strong et al. (2019) for COI, 12S, 16S, and 28S. For H3, amplification using Bioline Biolase taq (BIO-21042) was performed according to manufacturer’s instructions but modified to a 10uL reaction volume. Cycling parameters followed an initial denaturation at 94 °C for 3 min, followed by 35 cycles of denaturation at 94 °C for 35 s, annealing at 50 °C for 60 s and extension at 72 °C for 75 s, followed by a final extension at 72 °C for 10 min. The primers are listed in Table 3.
| Gene | Primer | Sequence (5′–3′) | Direction | Reference | Amplicon length |
|---|---|---|---|---|---|
| COI | jgLCO1490 | TIT CIA CIA AYC AYA ARG AYA TTG G | F | Geller et al. (2013) | 676 |
| COI | C1-N-2191R | CCC GGT AAA ATT AAA ATA TAA CTT C | R | Simon et al. (1994) | |
| 16S | 16Sa-L | TGC CTG TTT ATC AAA AAC AT | F | Palumbi (1996) | ∼510 |
| 16S | 16Sb-H2 | CTC CGG TTT GAA CTC AGA TCA | R | Palumbi (1996) | |
| 12S | 12SI | TGC CAG CAG YCG CGG TTA | F | Puillandre et al. (2009) | ∼545 |
| 12S | 12SIII | AGA GYG RCG GGC GAT GTG T | R | Puillandre et al. (2009) | |
| H3 | H3aF | ATG GCT CGT ACC AAG CAG ACV GC | F | Colgan et al. (1998) | 328 |
| H3 | H3aR | ATA TCC TTR GGC ATR ATR GTG AC | R | Colgan et al. (1998) | |
| 28S | C1 | ACC CGC TGA ATT TAA GCA T | F | Jovelin & Justine (2001) | ∼771 |
| 28S | D2 | TCC GTG TTT CAA GAC GGG | R | Jovelin & Justine (2001) |
All loci were sequenced in both directions; chromatograms were assembled and edited with Geneious Prime (Biomatters). See Table 1 for GenBank registration numbers.
Alignment and Phylogenetic analyses
Sequences for each locus were aligned separately with MUSCLE (Edgar, 2004) using default parameters as implemented in Geneious Prime. The final aligned length for each partition was as follows: COI–658 bp; 12S–613; 16S–547 bp; H3–328; 28S–787.
Phylogenetic analyses were conducted using maximum likelihood (ML) and Bayesian inference (BI) on the concatenated dataset. Maximum likelihood analyses were conducted in IQ-TREE ver. 1.6.12 (Nguyen et al., 2015) as implemented on the IQ-TREE web server (Trifinopoulos et al., 2016). The best-fit partitioning scheme and the most appropriate substitution model for each partition were estimated using ModelFinder (Kalyaanamoorthy et al., 2017) and partition models (Chernomor, von Haeseler & Minh, 2016). The best-fit partitioning scheme used distinct models for each locus, with the best-fit models as follows: GY+F+G4 (COI), TVM+F+I+G4 (12S), GTR+F+R5 (16S), MGK+F3X4+G4 (H3), GTR+F+I+G4 (28S). Nodal support was estimated with 1,000 ultrafast bootstrap replicates; nodes with values lower than 95% were considered unsupported (Hoang et al., 2018). For BI, the best fit models were determined with PartitionFinder 1.1.1 (Guindon et al., 2010; Lanfear et al., 2012; Lanfear et al., 2016), which favored GTR+I+G for each locus. Analyses were conducted using MrBayes 3.2.6 (Ronquist & Huelsenbeck, 2003) as implemented on the CIPRES Science Gateway (Miller, Pfeiffer & Schwartz, 2010), and consisted of two independent replicates with four heated chains each (temperature 0.02), and three swaps per swapping cycle, and were run for 50,000,000 Markov chain Monte Carlo (MCMC) generations with a sampling frequency of one tree every 1,000 generations. The first 25% were discarded as burn-in. Tracer 1.6 (Rambaut et al., 2014) was used to assess MCMC convergence and to ensure that all Effective Sample Size (ESS) values exceeded 200. A majority rule consensus tree was inferred with the sumt command. Nodal support was assessed with posterior probability (PP) of each node; nodes with PP lower than 90% were considered unsupported, PP = 90–95% moderately supported, and PP >95% highly supported.
Results
As indicated on the resulting consensus trees (Figs. 4 and 5), monophyly of Columbellidae is strongly supported to the exclusion of two species traditionally placed in the family. The monotypic Pseudanachis basedowi (Hedley, 1918) (Fig. 1L), was excluded from the ingroup in both the ML and Bayesian analyses, and grouped with Tomlinia with strong support in the Bayesian analysis (Fig. 4), suggesting that it may be a member of the new nassariid subfamily Tomliniinae Kantor et al. (2021). Pseudanachis basedowi is an Australian/ Indonesian species with a unique radula (Fig. 3E) that does not resemble those of Columbellidae. Placement of this species in Columbellidae has always been tenuous (e.g., Boss & Bieler, 1991; Wilson, 1998). In preliminary analyses of the multilocus dataset with a larger representation of buccinoidean outgroups (results not shown), another species formerly placed in the Columbellidae, Parviterebra brazieri (Angas, 1875), was excluded from the superfamily; subsequent analyses indicated it is a member of the Mangeliidae (Conoidea) (N. Puillandre, MNHN, pers. comm., 2022). Additional analyses are necessary to further refine the phylogenetic affinities of these taxa.
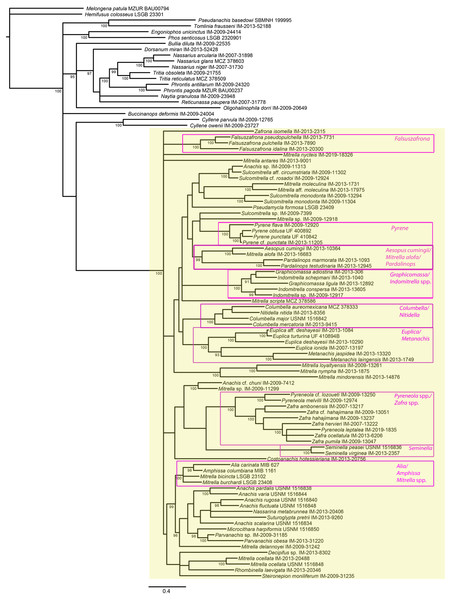
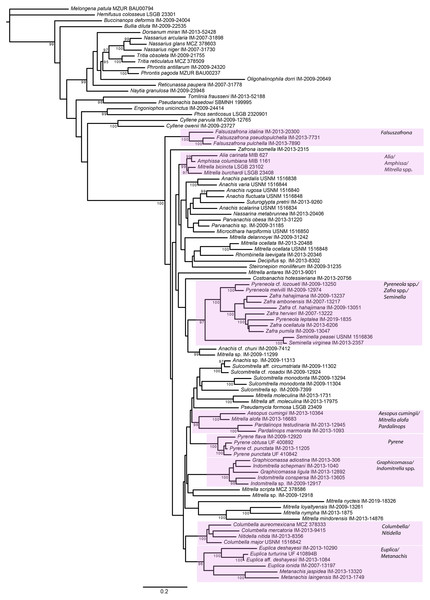
Figure 4: Phylogeny of Columbellidae obtained with Bayesian inference using a concatenated alignment of partial COI, 12S, 16S, 28S and H3 sequences.
Species traditionally placed in Columbellidae are indicated by the larger shaded box, and generic and supra-generic groupings discussed in the text are indicated by the outlined boxes. Posterior probabilities >90% are shown at the nodes.Figure 5: Phylogeny of Columbellidae obtained with maximum likelihood using a concatenated alignment of partial COI, 12S, 16S, 28S and H3 sequences.
Generic and supra-generic groupings are indicated by shaded boxes. Ultrafast bootstrap support values >95% are shown at the nodes.Within Columbellidae, interior nodes are almost uniformly without robust support in both the Bayesian and ML analyses. Data exploration varying the number of incomplete terminals demonstrated that missing data is not the cause (Fig. S1), nor is data conflict the source. Analyses of the individual gene trees and of the concatenated nuclear and mitochondrial gene datasets separately (Figs. S2, S3) revealed only isolated instances where strongly supported nodes conflicted, and these were only for relationships within or between genera (e.g., Graphicomassa and Indomitrella). Overall, more nodes were robustly resolved by mitochondrial than by nuclear genes, but both lack resolving power at deeper levels within the phylogeny. Indeed, the nuclear gene dataset was not capable of resolving the monophyly of the family when analyzed separately, with Tomlinia falling within the ingroup (Fig. S3).
Thus, from a taxonomic point of view, resolution within Columbellidae does not support any major clades that could be called subfamilies (Figs. 4 and 5). There are however several larger groupings without support, suggesting that future research with additional taxon sampling may offer some improvement. At present, the resulting trees support only the monophyly of several well-recognized genera and suprageneric groupings (Figs. 4 and 5). This indicates that combinations of shell and radular characters can be informative, however shell characters overall are highly homoplastic. Among these groups, some of the more noteworthy are those consisting of the included species of:
Aesopus cumingii + Mitrella alofa;
Columbella + Nitidella;
Euplica + Metanachis;
Falsuszafrona;
Graphicomassa + Indomitrella;
Pardalinops;
Pyrene;
Pyreneola + Zafra;
Seminella.
For the majority of these genera (except Aesopus, Indomitrella, Pyreneola and Zafra), a representative of the type species is included in the analysis.
A number of nominal columbellid genera are not supported as monophyletic. Notably, monophyly of the very often-used catch-all genera Mitrella and Anachis is not supported. Not surprisingly, these are also the most speciose nominal columbellid genera in MolluscaBase, with over 350 species between them. In our results, fourteen species of Mitrella are scattered across eight or more clades throughout the tree. The type species, Mitrella scripta (Linnaeus, 1758), groups with one other species in the ML analysis but without support, but otherwise does not appear to be related to the other species of Mitrella included in the analysis. Seven species of Anachis are grouped in four different clades, though five of these species, including the type species Anachis scalarina (Sowerby I, 1832) are part of a large, poorly supported suprageneric group in both trees.
Discussion
This is the first molecular analysis of the family Columbellidae, and it supports the monophyly of the family and confirms the utility of the unusual radular morphology found in this group as an important diagnostic character complex for the family. One genus and species traditionally included in Columbellidae falls outside the family, Pseudanachis basedowi (Hedley, 1918), which groups with Tomlinia frausseni of the family Nassariidae. This species is the only member of its genus, and it has a radular morphology that is atypical for Columbellidae (Fig. 3E), but its shell size and form is otherwise similar.
Results of Kantor et al.’s (2021) analysis of Buccinoidea suggest that Columbellidae is closely related to, if not derived within, the Nassariidae as currently conceived, in particular to members of the subfamily Cylleninae. However, this placement is without nodal support and there is no evident unambiguous synapomorphy that would support this grouping. Features that columbellids have in common with cyllenines include the small shell (usually 20 mm or less in length) and the (typically) smooth protoconch. The radula in columbellids and cyllenines is not similar; the rachidian in cyllenines has a series of pointed cusps whereas the columbellid center plate is acuspate; the lateral teeth have a single curved cusp whereas columbellids have secondary cusps. It may be that the distinctive columbellid radula has evolved from such a morphology, but no intermediate forms have so far been found. Kantor et al. (2021) considered it premature to revise the family classification of nassariids given the lack of support, other than to exclude the Buccinanopsinae. The monophyly of columbellids (excluding Pseudanachis) is not in doubt, but their position and rank, as well as the taxonomic fate of the remaining members of the Nassariidae, remain uncertain and require further analysis.
Within Columbellidae, resolution among interior nodes is problematic, however a number of genera and suprageneric groupings appear to have preliminary support pending inclusion of additional species. Typically, these groups as they are traditionally known consist of relatively few species with recognizable morphological characteristics and a strong regional identity. Among the genera that are supported as monophyletic or potentially so are the tropical Atlantic/ Eastern Pacific Columbella (including the monotypic genus Nitidella), the Indo-Pacific genera Euplica Dall, 1889, Metanachis Thiele, 1924, Pardalinops deMaintenon 2008, Pyrene, Pyreneola, Seminella, and Zafra. Columbella, which typifies the nominotypical subfamily in Radwin’s arrangement, is represented in our analysis by the type species C. mercatoria (Linnaeus, 1758) (Fig. 1A) and two others from the Eastern Pacific. Nested within Columbella in these results is also the monotypic Nitidella nitida (Lamarck, 1822) (Fig. 1B). The latter has a very different shell morphology (Columbella species are strombiform with a striated periostracum, but Nitidella is oliviform, glossy, and lacking the periostracum; both have weak early spire axials and weak columellar plaits). The radular tooth morphology, illustrated by Radwin (1977a for Columbella mercatoria, C. rusticoides and Nitidella nitida) and deMaintenon (1999 for Columbella major (as C. strombiformis) and N. nitida) is essentially identical and is also similar for other known Columbella species.
Euplica plus Metanachis form a clade, including their generic types, Euplica turturina (Lamarck, 1822) and Metanachis jaspidea (Sowerby I, 1844) (Figs. 1C, 1D). This is not surprising given that their radular morphology, illustrated by Sleurs (1985; Euplica varians, Metanachis jaspidea, M. laingensis) and by deMaintenon (2004; Euplica scripta, E. varians), is very distinct and more or less identical (Fig. 3E). During the present study, radulae were also examined (although not photographed) for Euplica deshayesii, MNHN IM-2013-10290, E. ionida MNHN IM-2007-13197, and Metanachis laingensis MNHN IM-2009-13022. These were all very similar but varied in lateral tooth length as would be expected for individuals of varying size. The shells on the other hand differ, with strombiform shells in Euplica and biconic shells in Metanachis. Members of both genera have weak to pronounced columellar plicae.
Aesopus cumingii (Reeve, 1859) also groups with Mitrella alofa (Hedley, 1899), and these two species form a well-supported clade together with the two species of Pardalinops deMaintenon 2008 (Figs. 1F–1H). Mitrella alofa is actually very similar to A. cumingii, but with a somewhat wider shell and larger aperture. Those two species and Pardalinops share no obvious morphological synapomorphies however. Their radulae have not been illustrated except for that of Pardalina testudinaria by Cernohorsky (1972). Aesopus is high spired with a sculptured shell (Fig. 1F), and Pardalinops has an unsculptured, squat biconic shell (Fig. 1H). The included species of Graphicomassa Iredale, 1929 (e.g., Fig. 1I) also form a monophyletic group with the included representatives of Indomitrella Oostingh, 1940. These also share no known synapomorphies and the radulae for Indomitrella species are unknown. The result calls into question the monophyly of both genera, insomuch as the species are intermixed. Analysis of additional taxa will help clarify the membership of these taxa.
The analyzed representatives of the genus Pyrene also comprise a well-supported clade, including the type species P. punctata (Bruguière, 1789) (Fig. 1E). The eight species of Pyrene are relatively large (15 to 25 mm long), biconic to pupoid, and unsculptured except for a weak to pronounced subsutural cord on the earliest teleoconch whorls. Radular morphology in these species, as shown by Radwin (1978) for P. punctata, is of a common form for the family and is not particularly informative.
Our analyses also indicate the newly named genus Falsuszafrona Pelorce, 2020 to be monophyletic. The included species had previously been placed in Zafrona Iredale, 1916. This was a questionable placement however, given that the 11 species of Falsuszafrona are restricted to the American tropics (e.g., Fig. 2I), whereas Zafrona, typified by Z. isomella (Duclos, 1840) (Fig. 2H), is a much smaller Indo-Pacific species with a very different radula. Falsuszafrona species have radulae similar to those of Euplica (as shown by Radwin, 1977a), but the radula of Zafrona isomella (as shown by Sleurs, 1987, as Z. nebulosa) is much reduced, with only two secondary lateral cusps and no center plate. The positions of Zafrona and Falsuszafrona are unresolved in the present analyses, but the results suggest that either or potentially both together could be sister to all other columbellids.
Another well-supported group consists of representative species of Zafra and Pyreneola with their respective type species (Figs. 2J, 2L). This is an interesting grouping because it consists of some of the truly tiny columbellid species of the Indo-Pacific, and the vast majority of the taxa in these groups have adult sizes below 4.5 mm. Shells in these genera are narrowly biconic and typically axially sculptured, with a restricted aperture and in some species a columellar plica. These species also are characterized by the lack of an operculum, which is generally present in other members of the family. The members of Seminella (Fig. 2K) also tend to group with these, with less support; they too are characterized by their tiny adult size, axial sculpture, lack of an operculum, and similar shell shape to Zafra and Pyreneola. The radulae in these three genera (Sleurs, 1987) are of a basic form common throughout the Columbellidae.
Finally, one more group that appears as a well-supported clade in both trees is a group comprised of Alia carinata, Amphissa columbiana, and two Mitrella species, M. burchardi and M. bicincta. This assemblage is interesting because these comprise all of the cold-water North Pacific species included in this analysis. So although this is morphologically an odd combination of both large and small species, with biconic, sculptured and unsculptured shells, geographically it makes sense.
One nominal genus that remains unresolved is Mitrella. This is problematic because it is easily the most speciose living nominal columbellid genus, with 220 valid extant species (MolluscaBase, 2021). Our phylogeny includes twelve individuals representing ten nominal species (e.g., Figs. 1J, 1K), which are distributed in seven clades across the entire phylogeny. One of these clades contains three tall-spired Indo-Pacific species (Mitrella loyaltyensis, M. nympha and M. mindorensis (Fig. 1J)), suggesting that at least that group may remain intact, but the others tend to group with members of other genera if anything. The relationships of the type species, Mitrella scripta (Linnaeus, 1758) from the Mediterranean, are essentially unresolved. Mitrella has always been a ‘wastebasket’ taxon (Plotnick & Wagner, 2006) as it lacks any synapomorphic diagnostic characters (species differ primarily in size, shape, inflation of the whorls, proportions and often color or pattern), and the radula is usually of the common form found throughout the family (e.g., Figs. 3A, 3C, 3D). Increased taxon sampling should help to resolve the membership of this genus and the unrelated species assigned to it.
Anachis is another genus that tends to accumulate new species without good reason; it currently consists of 102 valid extant species worldwide (MolluscaBase, 2021). Anachis was named by Adams & Adams (1853) to include fusiform species with axial ribs, but they did not indicate a type species; Anachis scalarina (Sowerby I, 1832) was later so designated by Tate (1868). Anachis scalarina in our results is also in an unresolved position. It tends to group with other tropical American species, albeit mostly without support, with representatives of a number of genera with axially and/or spirally sculptured biconic shells, including species of Anachis (e.g., Fig. 2A), Nassarina (e.g., Fig. 2D), Steironepion (e.g., Fig. 2E), Suturoglypta (e.g., Fig. 2F), Parvanachis and Microcithara. Radulae in these taxa vary (as shown by Radwin, 1977b, Radwin), and may prove to be taxonomically informative given further study.
The lack of support for suprageneric relationships is somewhat surprising given the utility of the targeted loci for resolving relationships among and within neogastropod and other caenogastropod families (e.g., Galindo et al., 2016; Fedosov et al., 2015; Fedosov et al., 2020; Kantor et al., 2017; Kantor et al., 2021; Strong et al., 2019). Exploratory analyses varying the quantity of missing data and of the nuclear and mitochondrial datasets separately (Fig. S1 through Fig. S3) showed that the low support values and lack of resolution is not a consequence of missing data. Somewhat counterintuitively, given the expectation that nuclear genes should be more conservative for better resolving interior nodes, the resolving power of both datasets is concentrated at the tips. This lack of resolution may be the result of Columbellidae’s comparatively recent origin and possibly rapid diversification relative to other neogastropod families, most of which have been present since the Cretaceous (Taylor, Morris & Taylor, 1980). The Columbellidae appears to be entirely Cenozoic (Taylor, Morris & Taylor, 1980; Squires, 2015), and underwent ‘great diversification’ from Miocene to Recent (Woodring, 1964; Squires, 2015). The greatest historical diversity is undoubtedly Recent. There is a ‘meager’ Paleogene fossil record consisting of a few species mostly assigned to the genera Astyris and Mitrella (Squires, 2015). Research on Miocene marine faunas has reported a few more diverse columbellid assemblages. For example, Woodring (1964) documented six genera and 15 species of columbellids (including four species each of Mitrella and Anachis, and six species of Strombina) from the Mid-Miocene Gatun Formation in the Panama Canal Zone, and Harzhauser & Landau (2021) found 37 species (including 12 species placed in Mitrella) in 15 genera from Mid-Miocene Central Paratethys deposits in Europe. Over half of the nominal genera documented in these works are modern; but given we have not so far found any conchological characters to be robustly informative for delineating most columbellid genera, the placement of many of these species and their relationships to modern forms cannot be confirmed, and such confirmation may indeed never be possible. One exception may be the 98 species in five nominal genera of the Neogene Strombina-group (Jung, 1989), which is undoubtedly the most noteworthy group of fossil columbellids. They are speciose, geographically well-circumscribed, and morphologically quite distinct (unusually large on average for columbellids, with a narrow strombiform shell and a high spire, and in some, a dorsal hump on the body whorl), so more likely to be monophyletic. Most of the present 70+ extant nominal columbellid genera do not appear to have a fossil record. Increased taxonomic sampling and a phylogenomic approach will be required to robustly resolve the deeper branches of the columbellid tree.
Conclusion
The objective of the present study was to evaluate the monophyly and relationships of Columbellidae using molecular data, and to clarify the relationships of nominal genera within the family. Reconstruction of the relationships within this group will provide a basis for investigating the evolution of a number of morphological and ecological characteristics that have long been of interest to marine gastropod researchers. The results of this study indicate that Columbellidae is monophyletic with the exception of two species (with an atypical radula morphology in one and an unknown radular morphology in the other), and thus the radular morphology used to define membership of the family is indeed unique to it. Relationships within the family are not however well enough resolved in our results to suggest a subfamilial or generic level classification, though several morphologically distinct genera and suprageneric groupings, most of which have similar radular or conchological characteristics, do appear to be monophyletic. Radular morphology appears to be more conservative in columbellids than shell form, and in some genera appears to be taxonomically informative. The two most nominally diverse genera, Mitrella and Anachis, are not monophyletic. Unfortunately, given Mitrella is also probably the most speciose fossil genus, resolving the relationships of fossil columbellids may prove to be extremely difficult. Future efforts to resolve the relationships within this group may benefit from increased taxon sampling to target groups not represented here, particularly of temperate lineages and the Strombina group, and broader genomic sampling potentially including mitogenomics.
Supplemental Information
Concatenated sequences used in phylogenetic analysis, fasta format
Phylogeny of Columbellidae obtained with maximum likelihood for a reduced taxonomic data set omitting terminals lacking more than one gene sequence
Phylogeny of Columbellidae obtained with maximum likelihood using a concatenated alignment of partial COI, 12S, 16S, 28S and H3 sequences for a reduced taxonomic subset omitting terminals lacking data for more than one locus. Ultrafast bootstrap support values >95% are shown at the nodes.
Phylogeny of Columbellidae obtained with maximum likelihood using mitochondrial gene sequences
Phylogeny of Columbellidae obtained with maximum likelihood using a concatenated alignment of mitochondrial gene (COI, 12S, 16S) sequences. Ultrafast bootstrap support values >95% are shown at the nodes.
Phylogeny of Columbellidae obtained with maximum likelihood using nuclear gene sequences
Phylogeny of Columbellidae obtained with maximum likelihood using a concatenated alignment of nuclear gene (28S and H3) sequences. Ultrafast bootstrap support values >95% are shown at the nodes.