Last week, we published ‘Strain- and plasmid-level deconvolution of a synthetic metagenome by sequencing proximity ligation products’, a very interesting study in which Hi-C—a technology originally designed for the study of 3D genome structure in eukaryotes—is used to measure the cellular co-localization of DNA sequences.
We invited the first author Christopher Beitel to comment on his research and his experience publishing with us.
PJ: Can you tell us a bit about yourself?

CB: I am a Ph.D. student in Jonathan Eisen’s lab at UC Davis. The two areas that I get most excited about are personalized medicine and open science. In the long term, I hope my current work on metagenomics brings us closer to more effective methods to profile individual human microbiomes. The methods I use are both computational and molecular and my training spans pure and applied mathematics, computing, genomics, and molecular biology. Outside the lab I am very interested in wellness and quantified self. I love running and meditation.
PJ: Can you briefly explain the research you published in PeerJ?
CB: Let’s say you have a trashcan full of shredded up phone books from five different cities in the US. And you want to put them back together. That would be easier if they were all printed on different colors of paper, right? In that case, the color of the paper is a signal of what was separate originally, before someone destroyed your strange collection of phonebooks. Once you’ve split the pieces of paper by color, then you can start taping them back together and although that’s a long road to travel, you’re in an easier position than if they were all printed on the same pasty grey paper.
In our study, we used Hi-C to create a signal for ourselves of what pieces of DNA came from the same cell. Hi-C is a proximity ligation method where we used formaldehyde to stick pieces of nearby DNA and protein together within intact cells. Then when we broke those cells open, cut up the DNA, and diluted everything. We followed that with ligation of free DNA ends. Basically the conditions created vastly favor the ligation of DNA fragments that were originally in the same cell.
In this experiment we intentionally constructed a very simple community to allow us to test the idea. And so far it works. We’ll see how this does on a more complex community.
PJ: What surprised you the most with these results?
CB: I was most surprised by the effectiveness of the MCL clustering algorithm. Some background, think of contigs like pieces of phonebooks. MCL is doing the job of looking at scraps of paper and grouping them by color. The surprising thing was that it did a nearly perfect job and up to that point we had not been able to cluster this well with any other clustering method. So naturally we spent a good amount of time trying to figure out how the result might be artifactual. Maybe we had unknowingly tipped off MCL to the right answer in the form of sorted input or in the way contigs were named. It appears this result is not an artifact. We did a pretty extensive exploration of the relevant parameter space and found some sensitivity but it appears robust on adequately filtered data.
The other surprising result of the paper was how plasmids tended to be associated with each other and with the chromosome of their host more frequently than anything else. I think that’s really cool.
PJ: Which figure in the manuscript do you think best summarizes your results?
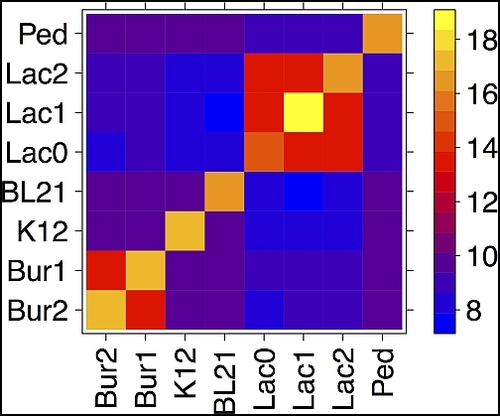
.
CB: Definitely Figure 2. This figure illustrates the signal I was talking about with that first question you asked me. That’s the one that shows you visually that replicons tend to be associated with themselves orders of magnitude more frequently than with others of the same cell. Then, in turn, those of the same cell tend to be associated with each other orders of magnitude more frequently with than those of different cells.
PJ: Why did you choose to reproduce the complete peer-review history of your article?
CB: Because making that information available could be beneficial to science and certainly does not do any harm. I don’t like the status quo system. A third possibility is post-publication review, something PeerJ could enable by simply allowing anyone with a PeerJ account to openly up- or down-vote a particular preprint or manuscript (or a particular aspect thereof).
PJ: How did you first hear about PeerJ, and what persuaded you to submit to us?
CB: You’re pretty well known. We initially came to PeerJ for the preprint server. That was a smooth experience. That together with the fact that you’re an open access journal, you have open review, and you don’t charge crazy fees for color figures.
PJ: How would you describe your experience of our submission / review process?
CB: Very nearly optimal.
PJ: Would you submit again, and would you recommend that your colleagues submit?
CB: Yes.
PJ: In conclusion, how would you describe PeerJ in three words?
CB: Refreshing snowflake laserbeam
PJ: Anything else you would like to talk about?
CB: If you support open access to publicly funded research products, show your support for California’s AB 609 (http://www.sparc.arl.org/advocacy/state/ab609), “California Taxpayer Access to Publicly Funded Research Legislation”.
Also I would love to see PeerJ article pages support dynamic SVG (d3.js) figures, and if you had a “fork this on GitHub” button for publications with software that would be pretty neat.
PJ: Many thanks for your time and your feedback!
CB: Thank you for chatting with me.
